Briefdefinitive Report
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TAXONOMIC STUDIES from RODENT OUTBREAK AREAS in the CHITTAGONG HILL TRACTS Nikhil
Bangladesh J. Zool. 46(2): 217-230, 2018 ISSN: 0304-9027 (print) 2408-8455 (online) NEW RECORDS OF RODENT SPECIES IN BANGLADESH: TAXONOMIC STUDIES FROM RODENT OUTBREAK AREAS IN THE CHITTAGONG HILL TRACTS Nikhil Chakma*, Noor Jahan Sarker, Steven Belmain1, Sohrab Uddin Sarker, Ken Aplin2 and Sontosh Kumar Sarker3 Department of Zoology, University of Dhaka, Dhaka-1000, Bangladesh Abstract: Rodents are regarded as crop pests, significant reservoirs and vectors for many zoonotic diseases around the world. Basic taxonomic information of rodents present in a locality can help understand which species are responsible as crop pest in that habitat. The phenomenon of the 50-year cycle of gregarious bamboo flowering and rodent outbreaks in the Chittagong Hill Tracts (CHT) of Bangladesh, rodents trapping were carried out in four habitats from March, 2009 to December, 2011 in Ruma upazila of Bandarban hill district. Variety of traps were used to capture small mammals. The captured species were measured and identified using taxonomical dichotomous keys and DNA bar-coding performed in Australia. A total of 14 different small mammalian species were captured of which nine belonging to the Muridae family, and one species each of Spalacidae, Sciuridae, Tupaiidae and Soricidae families. The dominant small mammal species captured were Rattus rattus (54.06%) followed by Mus musculus (26.39%), Rattus nitidus (10.98%), Suncus murinus (5.45%), Mus terricolor (1.09%), Mus cookii nagarum (0.97%), Cannomys badius (0.16%), Leopoldamys edwardsi (0.12%), Berylmys bowersi (0.12%), Vernaya fulva (0.08%), Rattus andamanensis (0.08%), Tupaia glis (0.04%) and Callosciurus pygerythrus (0.04%). -

Micromys Minutus)
Acta Theriol (2013) 58:101–104 DOI 10.1007/s13364-012-0102-0 SHORT COMMUNICATION The origin of Swedish and Norwegian populations of the Eurasian harvest mouse (Micromys minutus) Lars Råberg & Jon Loman & Olof Hellgren & Jeroen van der Kooij & Kjell Isaksen & Roar Solheim Received: 7 May 2012 /Accepted: 17 September 2012 /Published online: 29 September 2012 # Mammal Research Institute, Polish Academy of Sciences, Białowieża, Poland 2012 Abstract The harvest mouse (Micromys minutus) occurs the Mediterranean, from France to Russia and northwards throughout most of continental Europe. There are also two to central Finland (Mitchell-Jones et al. 1999). Recently, isolated and recently discovered populations on the it has also been found to occur in Sweden and Norway. Scandinavian peninsula, in Sweden and Norway. Here, we In 1985, an isolated population was discovered in the investigate the origin of these populations through analyses province of Dalsland in western Sweden (Loman 1986), of mitochondrial DNA. We found that the two populations and during the last decade, this population has been on the Scandinavian peninsula have different mtDNA found to extend into the surrounding provinces as well haplotypes. A comparison of our haplotypes to published as into Norway. The known distribution in Norway is sequences from most of Europe showed that all Swedish and limited to a relatively small area close to the Swedish Norwegian haplotypes are most closely related to the border, in Eidskog, Hedmark (van der Kooij et al. 2001; haplotypes in harvest mice from Denmark. Hence, the two van der Kooij et al., unpublished data). In 2007, another populations seem to represent independent colonisations but population was discovered in the province of Skåne in originate from the same geographical area. -

A Phylogeographic Survey of the Pygmy Mouse Mus Minutoides in South Africa: Taxonomic and Karyotypic Inference from Cytochrome B Sequences of Museum Specimens
A Phylogeographic Survey of the Pygmy Mouse Mus minutoides in South Africa: Taxonomic and Karyotypic Inference from Cytochrome b Sequences of Museum Specimens Pascale Chevret1*, Terence J. Robinson2, Julie Perez3, Fre´de´ric Veyrunes3, Janice Britton-Davidian3 1 Laboratoire de Biome´trie et Biologie Evolutive, UMR CNRS 5558, Universite´ Lyon 1, Villeurbanne, France, 2 Evolutionary Genomics Group, Department of Botany and Zoology, University of Stellenbosch, Stellenbosch, South Africa, 3 Institut des Sciences de l’Evolution de Montpellier, UMR CNRS 5554, Universite´ Montpellier 2, Montpellier, France Abstract The African pygmy mice (Mus, subgenus Nannomys) are a group of small-sized rodents that occur widely throughout sub- Saharan Africa. Chromosomal diversity within this group is extensive and numerous studies have shown the karyotype to be a useful taxonomic marker. This is pertinent to Mus minutoides populations in South Africa where two different cytotypes (2n = 34, 2n = 18) and a modification of the sex determination system (due to the presence of a Y chromosome in some females) have been recorded. This chromosomal diversity is mirrored by mitochondrial DNA sequences that unambiguously discriminate among the various pygmy mouse species and, importantly, the different M. minutoides cytotypes. However, the geographic delimitation and taxonomy of pygmy mice populations in South Africa is poorly understood. To address this, tissue samples of M. minutoides were taken and analysed from specimens housed in six South African museum collections. Partial cytochrome b sequences (400 pb) were successfully amplified from 44% of the 154 samples processed. Two species were identified: M. indutus and M. minutoides. The sequences of the M. indutus samples provided two unexpected features: i) nuclear copies of the cytochrome b gene were detected in many specimens, and ii) the range of this species was found to extend considerably further south than is presently understood. -

An Experimental Study with Micromys Minutus
Bachelor’s degree in Biology Final dissertation project Análise da dieta de pequenos mamíferos: un traballo experimental con Micromys minutus Análisis de dieta de pequeños mamíferos: un trabajo experimental con Micromys minutus Diet analysis of small mammals: an experimental study with Micromys minutus Anxo Pensado Moreira July, 2018 Academic supervisor: Sergio Rodríguez Roiloa ABSTRACT The ongoing growth of the human population and human activities have caused a rapid loss of biodiversity. Species play an important role in the ecosystem functioning. Thus, in order to conserve them, detailed insight into their biology is crucial. The Harvest mouse (Micromys minutus) is an understudied species whose populations across the UK have undergone an apparent decline of the 71 % over the past 18 years. Furthermore, there seems to be little information available on the diet of this species, which can be essential to improve welfare guides for captive populations as well as for future conservation actions for wild populations. Thereby, the aim of the present study was to shed some light on the diet of a captive population of Micromys minutus at the Wildwood Trust by assessing their (1) dietary preferences, (2) potential sex differences, (3) feeding time patterns and (4) intake rates. In order to do so, 5 individuals of Harvest mice were presented with different feeds and as a result their consumptions over a period of 14 days were obtained. Posteriorly, data was analysed by means of variance analysis (ANOVA) performed with SPSS, showing significant differences in the food intake of Micromys minutus depending upon food type and feeding time effects. The study revealed that the mice preferred blackberries, canary seeds, dried meal worms, naked oats and safflowers over white and red millet, although males had a higher consumption of dried meal worms and naked oats, whilst females preferred safflowers. -

Increased Geographic Sampling Reveals Considerable New Genetic
Mammalian Biology 79 (2014) 24–35 Contents lists available at ScienceDirect Mammalian Biology jou rnal homepage: www.elsevier.com/locate/mambio Original Investigation Increased geographic sampling reveals considerable new genetic diversity in the morphologically conservative African Pygmy Mice (Genus Mus; Subgenus Nannomys) a,∗ a d a,b,c Jennifer Lamb , Sarah Downs , Seth Eiseb , Peter John Taylor a School of Life Sciences, New Biology Building, University of KwaZulu-Natal, University Road, Westville, KwaZulu-Natal 3630, South Africa b Department of Ecology and Resource Management, School of Environmental Sciences, University of Venda, Post Bag X5050, Thohoyandou 0950, South Africa c Core Team Member, Centre for Invasion Biology, Department of Botany and Zoology, Stellenbosch University, Post Bag X1, Matieland 7602, South Africa d University of Namibia, Windhoek, Namibia a r a t i b s c l e i n f o t r a c t Article history: African endemic pygmy mice (Genus Mus; sub-genus Nannomys) have considerable economic and public Received 7 March 2013 health significance, and some species exhibit novel sex determination systems, making accurate knowl- Accepted 19 August 2013 edge of their phylogenetics and distribution limits important. This phylogenetic study was based on the by Frank E. Zachos mitochondrial control region and cytochrome b gene, for which a substantial body of published data was Available online 13 September 2013 available. Study specimens were sourced from eight previously unsampled or poorly sampled countries, and include samples morphologically identified as Mus bufo, M. indutus, M. callewaerti, M. triton and M. Keywords: neavei. These analyses increase the known genetic diversity of Nannomys from 65 to 102 haplotypes; at Nannomys least 5 unassigned haplotypes are distinguished by potentially species-level cytochrome b genetic dis- Mus bufo tances. -

SHREWS Robert H
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln The aH ndbook: Prevention and Control of Wildlife Wildlife Damage Management, Internet Center for Damage 1-1-1994 SHREWS Robert H. Schmidt Utah State University, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/icwdmhandbook Part of the Environmental Sciences Commons Schmidt, Robert H., "SHREWS" (1994). The Handbook: Prevention and Control of Wildlife Damage. 55. http://digitalcommons.unl.edu/icwdmhandbook/55 This Article is brought to you for free and open access by the Wildlife Damage Management, Internet Center for at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in The aH ndbook: Prevention and Control of Wildlife Damage by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Robert H. Schmidt Assistant Professor Department of Fisheries and Wildlife SHREWS Utah State University Logan, Utah 84322-5210 Fig. 1. A masked shrew, Sorex cinereus Identification Damage Prevention and Fumigants The shrew is a small, mouse-sized Control Methods None are registered. mammal with an elongated snout, a Trapping dense fur of uniform color, small eyes, Exclusion and five clawed toes on each foot (Fig. Mouse trap (snap trap). Rodent-proof structures also exclude 1). Its skull, compared to that of shrews. Small box trap. rodents, is long, narrow, and lacks the zygomatic arch on the lateral side Cultural Methods Pit trap. characteristic of rodents. The teeth are Mowing may decrease preferred Shooting small, sharp, and commonly dark- tipped. Pigmentation on the tips of the habitat and food. Not practical. teeth is caused by deposition of iron in Repellents Other Methods the outer enamel. -

Isolation, Characterization, and Evolutionary Divergence of Mouse Rnase 6: Evidence for Unusual Evolution in Rodents
J Mol Evol (2004) 59:657–665 DOI: 10.1007/s00239-004-2657-0 Isolation, Characterization, and Evolutionary Divergence of Mouse RNase 6: Evidence for Unusual Evolution in Rodents Kimberly D. Dyer,1 Helene F. Rosenberg,1 Jianzhi Zhang2 1 Eosinophil Biology Section, Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA 2 Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor MI 48109, USA Received: 26 February 2004 / Accepted: 7 June 2004 [Reviewing Editor: Dr. Lauren Ancel Meyers] Abstract. The evolution of the ribonuclease A Key words: Ribonuclease — RNase A superfamily (RNase A) vertebrate-specific enzyme family is inter- — RNase k6 — RNase 6 — Host defense esting in that specific gene lineages appear to be responding to unique selective pressures in wildly diverse manners to generate proteins that are capable Introduction of reducing the infectivity of viruses, killing systemic pathogens, and inducing the growth of blood vessels all Ribonuclease 6 is a member of the ribonuclease A while maintaining the signature motifs of a ribonu- (RNase A) superfamily, which is believed to be the clease. In this paper, we present the DNA sequence and only enzyme family that is limited to vertebrates gene structure of Mus musculus RNase 6 and examine (Lander et al. 2001). This family has been well char- the expression pattern and enzymatic activity of the acterized structurally, with most members having recombinant protein. M. musculus RNase 6 has a eight cysteines as well as the lysine and the two his- limited expression pattern compared to human RNase tidines that form the catalytic triad (Beintema et al. -

A Behavioural Syndrome, but Less Evidence for a Relationship with Cognitive Traits in a Spatial Orientation Context Andrea C
Schuster et al. Frontiers in Zoology (2017) 14:19 DOI 10.1186/s12983-017-0204-2 RESEARCH Open Access A behavioural syndrome, but less evidence for a relationship with cognitive traits in a spatial orientation context Andrea C. Schuster*, Uwe Zimmermann, Carina Hauer and Katharina Foerster Abstract Background: Animals show consistent individual behavioural differences in many species. Further, behavioural traits (personality traits) form behavioural syndromes, characterised by correlations between different behaviours. Mechanisms maintaining these correlations could be constrained due to underlying relationships with cognitive traits. There is growing evidence for the non-independence of animal personality and general cognitive abilities in animals, but so far, studies on the direction of the relationship between them revealed contradictory results. Still, it is hypothesised that individuals may exhibit consistent learning and decision styles. Fast behavioural types (consistently bolder and more active individuals) are expected to show faster learning styles. Slow behavioural types in contrast are assumed to learn slower but more accurately. This can be caused by a speed-accuracy trade-off that individuals face during decision making. We measured the repeatability of three personality and four spatial cognitive traits in adult Eurasian harvest mice (Micromys minutus). We analysed correlations among personality traits (behavioural syndrome). We further investigated the relationships between personality and spatial cognitive traits as a first step exploring the potential connection between personality and cognition in this species. Results: Our results showed that exploration, activity and boldness were repeatable in adult mice. Spatial recognition measured in a Y Maze was also significantly repeatable, as well as spatial learning performance and decision speed. -
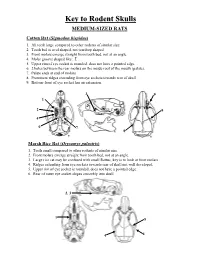
Key to Rodent Skulls MEDIUM-SIZED RATS Cotton Rat (Sigmodon Hispidus) 1
Key to Rodent Skulls MEDIUM-SIZED RATS Cotton Rat (Sigmodon hispidus) 1. All teeth large compared to other rodents of similar size. 2. Tooth bed is oval shaped, not teardrop shaped. 3. Front molars emerge straight from tooth bed, not at an angle. 4. Molar groove shaped like: Σ 5. Upper rim of eye socket is rounded, does not have a pointed edge. 6. 2 holes between the rear molars on the inside roof of the mouth (palate). 7. Palate ends at end of molars. 8. Prominent ridges extending from eye sockets towards rear of skull. 9. Bottom-front of eye socket has an extension. 3 9 1 2 5 4 6 7 8 Marsh Rice Rat (Orysomys palustris) 1. Teeth small compared to other rodents of similar size. 2. Front molars emerge straight from tooth bed, not at an angle. 3. Large rice rat may be confused with small Rattus, key is to look at front molars 4. Ridges extending from eye sockets towards rear of skull not well developed. 5. Upper rim of eye socket is rounded, does not have a pointed edge. 6. Rear of outer eye socket slopes smoothly into skull. 2, 3 1 5 4 6 LARGE-SIZED RATS Roof Rat (Rattus rattus) • Skull longer and pointer than cotton rat or rice rat. • Skull generally longer than 25 mm. 3. Front molars (and their tooth beds) strongly angled up towards front of skull. 4. Front molars large. 5. Tooth bed teardrop shaped, pointed towards rear. 6. Upper rim of eye sockets have a slight point into the sockets. -

Phylogeny of the Genus Apodemus with a Special Emphasis on the Subgenus Sylvaemus Using the Nuclear IRBP Gene and Two Mitochondrial Markers: Cytochrome B and 12S Rrna
MOLECULAR PHYLOGENETICS AND EVOLUTION Molecular Phylogenetics and Evolution 23 (2002) 123–136 www.academicpress.com Phylogeny of the genus Apodemus with a special emphasis on the subgenus Sylvaemus using the nuclear IRBP gene and two mitochondrial markers: cytochrome b and 12S rRNA J.R. Michaux,a,b,* P. Chevret,b M.-G. Filippucci,c and M. Macholand a Unite de Recherches Zoogeographiques, Institut de Zoologie, Quai Van Beneden, 22, 4020 Liege, Belgium b Laboratoire de Paleontologie—cc064, Institut des Sciences de l’Evolution de Montpellier (UMR 5554-CNRS), UM II, Place E. Bataillon, 34095 Montpellier Cedex 05, France c Dipartimento di Biologia, Universita di Roma, ‘‘Tor Vergata’’ Via della Ricerca Scientifica, 00133 Rome, Italy d Institute of Animal Physiology and Genetics, Academy of Sciences of the Czech Republic, Veverı 97, 60200 Brno, Czech Republic Received 11 June 2001; received in revised form 8 November 2001 Abstract Phylogenetic relationships among 17 extant species of Murinae, with special reference to the genus Apodemus, were investigated using sequence data from the nuclear protein-coding gene IRBP (15 species) and the two mitochondrial genes cytochrome b and 12S rRNA (17 species). The analysis of the three genes does not resolve the relationships between Mus, Apodemus, and Rattus but separates Micromys from these three genera. The analysis of the two mitochondrial regions supported an association between Apodemus and Tokudaia and indicated that these two genera are more closely related to Mus than to Rattus or Micromys. Within Apodemus, the mitochondrial data sets indicated that 8 of the 9 species analyzed can be sorted into two main groups: an Apodemus group, with A. -

Karyotypes of the Japanese Harvest Mouse (Micromys Minutus Japonicus) from Fukuoka and the Tsushima Islands
九州大学学術情報リポジトリ Kyushu University Institutional Repository Karyotypes of the Japanese Harvest Mouse (Micromys minutus japonicus) from Fukuoka and the Tsushima Islands Okura, Nobuhiko Zoological Laboratory, Faculty of Agriculture, Kyushu University Shiraishi, Satoshi Zoological Laboratory, Faculty of Agriculture, Kyushu University Uchida, Teruaki Zoological Laboratory, Faculty of Agriculture, Kyushu University https://doi.org/10.5109/23787 出版情報:九州大学大学院農学研究院紀要. 28 (4), pp.177-183, 1984-03. 九州大学農学部 バージョン: 権利関係: J. Fat. Agr., Kyushu Univ., 28 (4), 177-183 (1984) Karyotypes of the Japanese Harvest Mouse (Micromys minutus iaponicus) from Fukuoka and the Tsushima Islands* Nobuhiko Okurat, Satoshi Shiraishi and Teru Aki Uchida Zoological Laboratory, Faculty of Agriculture, Kyushu University 46-06, Fukuoka 812 (Received November 29, 1983) Karyotypes of the Japanese harvest mouse, Micromys nzinutus japonicus, from two populations occurring in Fukuoka and the Tsushima Islands, were examined in detail using conventional, G- and C-band stainings. There was no difference in karyotype both between the two populations and within each population. The karyotypes in the two populations concerned were much the same as in other Japanese and Eurasian populations. Thus, it may be said that the harvest mouse has a quite conservative karyotype in spite of its wide geographical distribution. So far as it is based upon the karyotype, this conservatism does not support a specific division of the Japanese harvest mouse (M. japonicus) from the Eurasian one (M. minutus), which was proposed by Tokuda (1941), thus we regard the Japanese harvest mouse as M. minutus; moreover, M. minutus a&ii (Kuroda, 1922) from Tsushima seems to be a synonym of M. minutus japonicus (Thomas, 1905) from the mainland of Kyushu. -

Evolutionary and Dispersal History of Eurasian House Mice Mus Musculus Clarified by More Extensive Geographic Sampling of Mitochondrial DNA
Heredity (2013) 111, 375–390 & 2013 Macmillan Publishers Limited All rights reserved 0018-067X/13 www.nature.com/hdy ORIGINAL ARTICLE Evolutionary and dispersal history of Eurasian house mice Mus musculus clarified by more extensive geographic sampling of mitochondrial DNA H Suzuki1, M Nunome1, G Kinoshita1, KP Aplin2, P Vogel3, AP Kryukov4, M-L Jin5, S-H Han6, I Maryanto7, K Tsuchiya8, H Ikeda9, T Shiroishi10, H Yonekawa11 and K Moriwaki12 We examined the sequence variation of mitochondrial DNA control region and cytochrome b gene of the house mouse (Mus musculus sensu lato) drawn from ca. 200 localities, with 286 new samples drawn primarily from previously unsampled portions of their Eurasian distribution and with the objective of further clarifying evolutionary episodes of this species before and after the onset of human-mediated long-distance dispersals. Phylogenetic analysis of the expanded data detected five equally distinct clades, with geographic ranges of northern Eurasia (musculus, MUS), India and Southeast Asia (castaneus, CAS), Nepal (unspecified, NEP), western Europe (domesticus, DOM) and Yemen (gentilulus). Our results confirm previous suggestions of Southwestern Asia as the likely place of origin of M. musculus and the region of Iran, Afghanistan, Pakistan, and northern India, specifically as the ancestral homeland of CAS. The divergence of the subspecies lineages and of internal sublineage differentiation within CAS were estimated to be 0.37–0.47 and 0.14–0.23 million years ago (mya), respectively, assuming a split of M. musculus and Mus spretus at 1.7 mya. Of the four CAS sublineages detected, only one extends to eastern parts of India, Southeast Asia, Indonesia, Philippines, South China, Northeast China, Primorye, Sakhalin and Japan, implying a dramatic range expansion of CAS out of its homeland during an evolutionary short time, perhaps associated with the spread of agricultural practices.