Polyploidy Spectrum: a New Marker in Gut: First Published As 10.1136/Gutjnl-2018-318021 on 12 April 2019
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
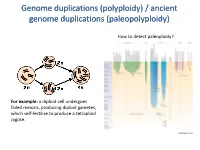
Polyploidy) / Ancient Genome Duplications (Paleopolyploidy
Genome duplications (polyploidy) / ancient genome duplications (paleopolyploidy) How to detect paleoploidy? For example: a diploid cell undergoes failed meiosis, producing diploid gametes, which self-fertilize to produce a tetraploid zygote. Timing of duplication by trees (phylogenetic timing) Phylogenetic timing of duplicates b Paramecium genome duplications Comparison of two scaffolds originating from a common ancestor at the recent WGD Saccharomyces cerevisiae Just before genome duplication Just after genome duplication More time after genome duplication Unaligned view (removing gaps just like in cerev has occurred) Saccharomyces cerevisiae Problem reciprocal gene loss (extreme case); how to solve? Problem reciprocal gene loss (extreme case); how to solve? Just before genome duplication Outgroup! Just after genome duplication Outgroup Just after genome duplication Outgroup More time after genome duplication Outgroup Problem (extreme case); how to solve? Outgroup Outgroup Outgroup Outgroup Outgroup Using other genomes Wong et al. 2002 PNAS Centromeres Vertebrate genome duplication Nature. 2011 Apr 10. [Epub ahead of print] Ancestral polyploidy in seed plants and angiosperms. Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, Depamphilis CW. Flowering plants Flowering MOSS Vertebrates Teleosts S. serevisiae and close relatives Paramecium Reconstructed map of genome duplications allows unprecedented mapping -

The Diversity of Plant Sex Chromosomes Highlighted Through Advances in Genome Sequencing
G C A T T A C G G C A T genes Review The Diversity of Plant Sex Chromosomes Highlighted through Advances in Genome Sequencing Sarah Carey 1,2 , Qingyi Yu 3,* and Alex Harkess 1,2,* 1 Department of Crop, Soil, and Environmental Sciences, Auburn University, Auburn, AL 36849, USA; [email protected] 2 HudsonAlpha Institute for Biotechnology, Huntsville, AL 35806, USA 3 Texas A&M AgriLife Research, Texas A&M University System, Dallas, TX 75252, USA * Correspondence: [email protected] (Q.Y.); [email protected] (A.H.) Abstract: For centuries, scientists have been intrigued by the origin of dioecy in plants, characterizing sex-specific development, uncovering cytological differences between the sexes, and developing theoretical models. Through the invention and continued improvements in genomic technologies, we have truly begun to unlock the genetic basis of dioecy in many species. Here we broadly review the advances in research on dioecy and sex chromosomes. We start by first discussing the early works that built the foundation for current studies and the advances in genome sequencing that have facilitated more-recent findings. We next discuss the analyses of sex chromosomes and sex-determination genes uncovered by genome sequencing. We synthesize these results to find some patterns are emerging, such as the role of duplications, the involvement of hormones in sex-determination, and support for the two-locus model for the origin of dioecy. Though across systems, there are also many novel insights into how sex chromosomes evolve, including different sex-determining genes and routes to suppressed recombination. We propose the future of research in plant sex chromosomes should involve interdisciplinary approaches, combining cutting-edge technologies with the classics Citation: Carey, S.; Yu, Q.; to unravel the patterns that can be found across the hundreds of independent origins. -

Cell Size, Polyploidy, and Sex Determination in Nasonia
Cell size, polyploidy, and sex determination in Nasonia Supervision by: Kelley Leung Contact: [email protected], room 5172.0662; language: English olyploidy is the condition of having more Pthan the usual number of chromosome sets. In many ways polyploidy is detrimental because of problems of infertility and abnormal cell biol- ogy. However, polyploidization has happened many times via whole genome duplication in the evolutionary tree (including for humans!). It may have provided benefits such as addition- al gene copies and increased hardiness. This begs the question, how do polyploids overcome disadvantages to derive benefits? One possible adaptation that has allowed polyploids to persist is cell reduction mech- anisms. Polyploid cells are larger than normal cells. In vertebrate polyploids, cell re- duction mechanisms are used to retain fairly normal overall body size and physiology. However, it is unknown if this also holds for invertebrate polyploids, including insects. We will use the parasitoid wasp system Nasonia vitripennis to investigate whether there are cell reduction mechanisms in insect polyploids. Like all hymenopterans, N. vitripen- nis has a haplodiploid sex determination system: normally, unfertilized eggs become haploid (1n) males and fertilized eggs become diploid (2n) females. However, there are many ways that ploidy levels change. Some are naturally occurring. Others can be created by using RNAi knockdown to obtain null mutants of various sex determination genes. These lines produce haploid (1n) females, diploid males -

Homoeologous Shuffling and Chromosome Compensation
Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus Zhiyong Xiong, Robert T. Gaeta, and J. Chris Pires1 Division of Biological Sciences, University of Missouri, Columbia, MO 65211 Edited by Susan R. Wessler, University of Georgia, Athens, GA, and approved March 31, 2011 (received for review September 22, 2010) Polyploidy has contributed to the evolution of eukaryotes, partic- B. napus affords an excellent opportunity for conducting cy- ularly flowering plants. The genomic consequences of polyploidy togenetic investigations of evolution in a resynthesized allo- have been extensively studied, but the mechanisms for chromo- polyploid. Natural B. napus (AACC; 2n = 38) is thought to have some stability and diploidization in polyploids remain largely un- formed 5,000 to 10,000 y ago by the hybridization of ancestors of known. By using new cytogenetic tools to identify all of the Brassica rapa (AA; 2n = 20) and Brassica oleracea (CC; 2n = 18) homoeologous chromosomes, we conducted a cytological investi- (28, 29, 30). B. rapa and B. oleracea are also ancient polyploids, gation of 50 resynthesized Brassica napus allopolyploids across gen- and large-scale chromosome rearrangements occurred in the A erations S0:1 to S5:6 and in the S10:11 generation. Changes in copy and C genomes following divergence from a common ancestor number of individual chromosomes were detected in the S0:1 gen- (31). Several studies have demonstrated that genetic changes eration and increased in subsequent generations, despite the fact caused by homoeologous chromosome rearrangement are com- that the mean chromosome number among lines was approxi- mon in newly resynthesized B. -

Multiple Large-Scale Gene and Genome Duplications During the Evolution of Hexapods
Multiple large-scale gene and genome duplications during the evolution of hexapods Zheng Lia,1, George P. Tileyb,c,1, Sally R. Galuskaa, Chris R. Reardona, Thomas I. Kiddera, Rebecca J. Rundella,d, and Michael S. Barkera,2 aDepartment of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ 85721; bDepartment of Biology, University of Florida, Gainesville, FL 32611; cDepartment of Biology, Duke University, Durham, NC 27708; and dDepartment of Environmental and Forest Biology, State University of New York College of Environmental Science and Forestry, Syracuse, NY 13210 Edited by Michael Freeling, University of California, Berkeley, CA, and approved March 12, 2018 (received for review June 14, 2017) Polyploidy or whole genome duplication (WGD) is a major contrib- than 800,000 described hexapod species (25) are known polyploids utor to genome evolution and diversity. Although polyploidy is (17, 20). However, until recently there were limited data available recognized as an important component of plant evolution, it is to search for evidence of paleopolyploidy among the hexapods generally considered to play a relatively minor role in animal and other animal clades. Thus, the contributions of polyploidy to evolution. Ancient polyploidy is found in the ancestry of some animal evolution and the differences with plant evolution have animals, especially fishes, but there is little evidence for ancient remained unclear. WGDs in other metazoan lineages. Here we use recently published To search for evidence of WGDs among the hexapods, we transcriptomes and genomes from more than 150 species across the leveraged recently released genomic data for the insects (26). insect phylogeny to investigate whether ancient WGDs occurred Combined with additional datasets from public databases, we assembled 128 transcriptomes and 27 genomes with at least one during the evolution of Hexapoda, the most diverse clade of animals. -

The Frequency of Polyploid Speciation in Vascular Plants
The frequency of polyploid speciation in vascular plants Troy E. Wooda,b,1, Naoki Takebayashic, Michael S. Barkerb,d, Itay Mayrosee, Philip B. Greenspoond, and Loren H. Riesebergb,d aInstitute for Evolution and Biodiversity, University of Mu¨nster, 48149 Mu¨nster, Germany; bDepartment of Biology, Indiana University, Bloomington, IN 47405; cInstitute of Arctic Biology and Department of Biology and Wildlife, University of Alaska, Fairbanks, AK 99775; and Departments of dBotany and eZoology, University of British Columbia, Vancouver, BC, Canada V6T 1Z4 Edited by Peter R. Crane, University of Chicago, Chicago, IL, and approved June 23, 2009 (received for review November 13, 2008) Since its discovery in 1907, polyploidy has been recognized as an contrast, polyploid incidence is distributed less equitably among 2 ϭ Ͻ 2 ϭ important phenomenon in vascular plants, and several lines of families ( 198 4,259.41, P 0.00001, R 0.116). Overall, with evidence indicate that most, if not all, plant species ultimately have the exception of the species-poor gymnosperms, vascular plant a polyploid ancestry. However, previous estimates of the fre- species derived from recent polyploid events are ubiquitous and quency of polyploid speciation suggest that the formation and represent a large fraction of named diversity, a pattern that also establishment of neopolyploid species is rare. By combining infor- holds for bryophytes (12). Interestingly, generic base counts mation from the botanical community’s vast cytogenetic and are negatively associated with polyploid incidence in angio- 2 ϭ Ͻ 2 ϭ phylogenetic databases, we establish that 15% of angiosperm and sperms ( 3 2,798.01, P 0.00001, R 0.085; Fig. -

Polyploidy in Drosophila Melanocaster With
POLYPLOIDY IN DROSOPHILAMELANOCASTER WITH TWO ATTACHED X CHROMOSOMES L. V. MORGAN Columbia University, New York City Received October 4, 1924 TABLE OF CONTENTS PAGE INTRODUCTION.................................................................. 148 Females with attached X’s. ...................................................... 150 Patroclinous females. ............................................................ 151 Breaking apart of the attached X’s. ............................................... 152 Triploid female.. ................................................................ 154 Crossing over in triploid females. ................................................. 160 Crossing over between the attached chromosomes. .................................. 162 Recessives showing mutant characters determined by differentials in the left end of the chromosome ................................................................. 162 Recessives showing mutant characters determined by differentials in the right end and middle of the chromosome.. .................................................... 163 Redistribution of differentials. .................................................... 165 Crossing over among three chromosomes. .......................................... 166 Recurrence of additional chromosomes. ............................................ 167 Tetraploid female. ............................................................... 167 Non-disjunction in males, ........................................................ 172 Composites .................................................................... -

Chromosome Polyploidization in Human Leukocytes Induced by Radiation
CHROMOSOME POLYPLOIDIZATION IN HUMAN LEUKOCYTES INDUCED BY RADIATION Carlos E. Nasjleti and Herbert H. Spencer The Veterans Administration Hospital and The University of Michigan, Ann Arbor, Mich. INTRODUCTION A great quantity of literature has accumulated on the effects of radiation and radiomimetic agents upon cells. Much of this work has been performed on mam- malian cells both in vivo and in tissue cultures. Some of these damaging effects can be seen in dividing cells. After the development of a technique for tissue culture of human peripheral blood, we were able to demonstrate abnormal patterns and chromosomal poly- ploidy in cultures from patients who had been treated with A-8103 (a piperazine derivative with antitumor activity) ,l nitrogen mustard, 6-mercaptopurine, and A-649 (another antitumor agent).2 During treatment and for some time after treatment in these patients, structural changes could be seen in the chromosomes as either chromatid or chromosome deletions and chromosomal breakage. Abnor- mally shaped chromosomes were also seen, particularly ring forms and dicentrics. Although aneuploidy was observed, the major numerical variation consisted of metaphases showing polyploidy with random distribution of chromosomes and diplochromosomes. When normal human leukocytes were tested in vitro with the chemotherapeutic agents already mentioned, as well as with streptonigrin and cycl~phosphamide,~similar results were obtained. From these studies we postu- lated that two different types of damage were induced in the cells by the chemicals. One type of damage is apparent in the hereditary material, and it can be charac- terized by the structural chromosomal changes. The second type of damage is interpreted as damage to the mitotic apparatus of the cells, as evidenced by the induction of polyploidy. -

The Evolutionary Consequences of Polyploidy
Leading Edge Review The Evolutionary Consequences of Polyploidy Sarah P. Otto1,* 1Department of Zoology, University of British Columbia, 6270 University Boulevard, Vancouver BC V6T 1Z4 Canada *Correspondence: [email protected] DOI 10.1016/j.cell.2007.10.022 Polyploidization, the addition of a complete set of chromosomes to the genome, represents one of the most dramatic mutations known to occur. Nevertheless, polyploidy is well toler- ated in many groups of eukaryotes. Indeed, the majority of flowering plants and vertebrates have descended from polyploid ancestors. This Review examines the short-term effects of polyploidization on cell size, body size, genomic stability, and gene expression and the long-term effects on rates of evolution. One of the most striking features of genome structure homology but are sufficiently distinct due to their sepa- is its lability. From small-scale rearrangements to large- rate origins, these pairs of chromosomes are referred scale changes in size, genome comparisons among to as homeologs (see Figure 1). Polyploidy is especially species reveal that variation is commonplace. Even over prevalent among hybrid taxa, an association thought the short time course of laboratory experiments, chro- to be driven by problems with meiotic pairing in diploid mosomal rearrangements, duplications/deletions of hybrids, which are solved if each homeologous chromo- chromosome segments, and shifts in ploidy have been some has its own pairing partner. Additionally, diploid observed and have contributed to adaptation (Dunham hybrids form unreduced gametes (which have the same et al., 2002; Gerstein et al., 2006; Riehle et al., 2001). number of chromosomes as somatic cells) at unusually Changes in genome structure typically have immediate high rates (Ramsey and Schemske, 2002), increasing the effects on the phenotype and fitness of an individual. -

Effect of Polyploidy Induction on Natural Metabolite Production in Medicinal Plants
biomolecules Review Effect of Polyploidy Induction on Natural Metabolite Production in Medicinal Plants Hadi Madani 1, Ainoa Escrich 2 , Bahman Hosseini 1, Raul Sanchez-Muñoz 3 , Abbas Khojasteh 4 and Javier Palazon 4,* 1 Department of Horticulture, Faculty of Agriculture, Urmia University, Urmia 5756151818, Iran; [email protected] (H.M.); [email protected] (B.H.) 2 Department of Experimental and Health Sciences, Universitat Pompeu Fabra, 08003 Barcelona, Spain; [email protected] 3 Laboratory of Functional Plant Biology, Department of Biology, Ghent University, K.L. Ledeganckststraat 35, B-9000 Ghent, Belgium; [email protected] 4 Department of Plant Physiology, Faculty of Pharmacy, University of Barcelona, Av. Joan, XXIII, 08028 Barcelona, Spain; [email protected] * Correspondence: [email protected] Abstract: Polyploidy plays an important role in plant diversification and speciation. The ploidy level of plants is associated with morphological and biochemical characteristics, and its modification has been used as a strategy to alter the quantitative and qualitative patterns of secondary metabolite production in different medicinal plants. Polyploidization can be induced by many anti-mitotic agents, among which colchicine, oryzalin, and trifluralin are the most common. Other variables involved in the induction process include the culture media, explant types, and exposure times. Due to the effects of polyploidization on plant growth and development, chromosome doubling has been Citation: Madani, H.; Escrich, A.; applied in plant breeding to increase the levels of target compounds and improve morphological Hosseini, B.; Sanchez-Muñoz, R.; characteristics. Prompted by the importance of herbal medicines and the increasing demand for Khojasteh, A.; Palazon, J. Effect of drugs based on plant secondary metabolites, this review presents an overview of how polyploidy Polyploidy Induction on Natural can be used to enhance metabolite production in medicinal plants. -

Polyploidy in Plants: Pharmacognosy
IOSR Journal Of Pharmacy And Biological Sciences (IOSR-JPBS) e-ISSN:2278-3008, p-ISSN:2319-7676. Volume 15, Issue 3 Ser. I (May –June 2020), PP 30-38 www.Iosrjournals.Org Polyploidy In Plants: Pharmacognosy Manorama Ratre* *School of Pharmacy,Chouksey Engineering College, Bilaspur, Chhattisgarh, India Abstract: Polyploidy or make greater in number of chromosomes have been found in living cells of plants as well as animals. Polyploidy is the strength of modern genetics or modern molecular technology in pharmacognosy to provide something new or combinations of characters in the field of plant breeding programs of medicinal plants and also for foods or crops. So the polyploidy is a perfect, popular and futuristic matter for review. Polyploidy is the most essential and useful because life on earth is present mainly, who depends on foods or crops and medicines for live their life. New significant for genome duplications should be measured with ancient ideas and new challenges of meiosis and mitosis. By the help of polyploidy we can be belief predicts which are crops and medicinal plants, usefully improved to obtained stable autopolyploids and which genomes or species adapted and merged to produce victorious new allopolyploids. This review gives information about the meaning of polyploidy, types, causes, factors which promotes polyploidy, examples, mechanisms, applications, advantages and disadvantages of polyploidy. By which we can predicts new significance for future prospective. Key words:- Polyploidy, genetics, molecular technology, plant -

Evidence That Polyploidy in Esophageal Adenocarcinoma Originates from Mitotic Slippage Caused by Defective Chromosome Attachments
Cell Death & Differentiation (2021) 28:2179–2193 https://doi.org/10.1038/s41418-021-00745-8 ARTICLE Evidence that polyploidy in esophageal adenocarcinoma originates from mitotic slippage caused by defective chromosome attachments 1 2 3 3 4 Stacey J. Scott ● Xiaodun Li ● Sriganesh Jammula ● Ginny Devonshire ● Catherine Lindon ● 2 1 Rebecca C. Fitzgerald ● Pier Paolo D’Avino Received: 2 July 2020 / Revised: 22 January 2021 / Accepted: 28 January 2021 / Published online: 1 March 2021 © The Author(s) 2021. This article is published with open access Abstract Polyploidy is present in many cancer types and is increasingly recognized as an important factor in promoting chromosomal instability, genome evolution, and heterogeneity in cancer cells. However, the mechanisms that trigger polyploidy in cancer cells are largely unknown. In this study, we investigated the origin of polyploidy in esophageal adenocarcinoma (EAC), a highly heterogenous cancer, using a combination of genomics and cell biology approaches in EAC cell lines, organoids, and tumors. We found the EAC cells and organoids present specific mitotic defects consistent with problems in the attachment of chromosomes to the microtubules of the mitotic spindle. Time-lapse analyses confirmed that EAC cells have problems in 1234567890();,: 1234567890();,: congressing and aligning their chromosomes, which can ultimately culminate in mitotic slippage and polyploidy. Furthermore, whole-genome sequencing, RNA-seq, and quantitative immunofluorescence analyses revealed alterations in the copy number, expression, and cellular distribution of several proteins known to be involved in the mechanics and regulation of chromosome dynamics during mitosis. Together, these results provide evidence that an imbalance in the amount of proteins implicated in the attachment of chromosomes to spindle microtubules is the molecular mechanism underlying mitotic slippage in EAC.