The Evolutionary Consequences of Polyploidy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Adaptation from Standing Genetic Variation and from Mutation
Adaptation from standing genetic variation and from mutation Experimental evolution of populations of Caenorhabditis elegans Sara Carvalho Dissertation presented to obtain the Ph.D degree in Biology Instituto de Tecnologia Química e Biológica | Universidade Nova de Lisboa Oeiras, Janeiro, 2012 Adaptation from standing genetic variation and from mutation Experimental evolution of populations of Caenorhabditis elegans Sara Carvalho Dissertation presented to obtain the Ph.D degree in Evolutionary Biology Instituto de Tecnologia Química e Biológica | Universidade Nova de Lisboa Research work coordinated by: Oeiras, Janeiro, 2012 To all the people I love. Table of contents List of Figures 3 List of Tables 5 Acknowledgements 7 Abstract 9 Resumo 13 CHAPTER 1 – Introduction 17 1.1 And yet…it changes 18 1.1.2 Evolution and adaptation 19 1.1.3 Mutation and standing genetic variation 25 Mutation 26 Standing genetic variation 32 Mutation versus standing genetic variation 34 Genetic recombination among adaptive alleles 35 1.1.4 Other players in evolution 37 1.1.5 Evolution in the wild and in the lab 40 1.1.6 Objectives 43 1.2 Caenorhabditis elegans as a model for experimental evolution 45 1.2.1 Experimental populations of C. elegans 50 1.3 References 53 CHAPTER 2 – Adaptation from high levels of standing genetic 63 variation under different mating systems 2.1 Summary 64 2.2 Introduction 64 2.3 Materials and Methods 69 2.4 Results 84 2.5 Discussion 98 2.6 Acknowledgements 103 2.7 References 103 2.8 Supplementary information 110 1 CHAPTER 3 – Evolution -

Human Chromosome‐Specific Aneuploidy Is Influenced by DNA
Article Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features Marie Dumont1,†, Riccardo Gamba1,†, Pierre Gestraud1,2,3, Sjoerd Klaasen4, Joseph T Worrall5, Sippe G De Vries6, Vincent Boudreau7, Catalina Salinas-Luypaert1, Paul S Maddox7, Susanne MA Lens6, Geert JPL Kops4 , Sarah E McClelland5, Karen H Miga8 & Daniele Fachinetti1,* Abstract Introduction Intrinsic genomic features of individual chromosomes can contri- Defects during cell division can lead to loss or gain of chromosomes bute to chromosome-specific aneuploidy. Centromeres are key in the daughter cells, a phenomenon called aneuploidy. This alters elements for the maintenance of chromosome segregation fidelity gene copy number and cell homeostasis, leading to genomic instabil- via a specialized chromatin marked by CENP-A wrapped by repeti- ity and pathological conditions including genetic diseases and various tive DNA. These long stretches of repetitive DNA vary in length types of cancers (Gordon et al, 2012; Santaguida & Amon, 2015). among human chromosomes. Using CENP-A genetic inactivation in While it is known that selection is a key process in maintaining aneu- human cells, we directly interrogate if differences in the centro- ploidy in cancer, a preceding mis-segregation event is required. It was mere length reflect the heterogeneity of centromeric DNA-depen- shown that chromosome-specific aneuploidy occurs under conditions dent features and whether this, in turn, affects the genesis of that compromise genome stability, such as treatments with micro- chromosome-specific aneuploidy. Using three distinct approaches, tubule poisons (Caria et al, 1996; Worrall et al, 2018), heterochro- we show that mis-segregation rates vary among different chromo- matin hypomethylation (Fauth & Scherthan, 1998), or following somes under conditions that compromise centromere function. -
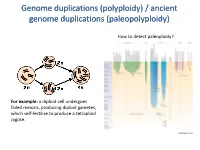
Polyploidy) / Ancient Genome Duplications (Paleopolyploidy
Genome duplications (polyploidy) / ancient genome duplications (paleopolyploidy) How to detect paleoploidy? For example: a diploid cell undergoes failed meiosis, producing diploid gametes, which self-fertilize to produce a tetraploid zygote. Timing of duplication by trees (phylogenetic timing) Phylogenetic timing of duplicates b Paramecium genome duplications Comparison of two scaffolds originating from a common ancestor at the recent WGD Saccharomyces cerevisiae Just before genome duplication Just after genome duplication More time after genome duplication Unaligned view (removing gaps just like in cerev has occurred) Saccharomyces cerevisiae Problem reciprocal gene loss (extreme case); how to solve? Problem reciprocal gene loss (extreme case); how to solve? Just before genome duplication Outgroup! Just after genome duplication Outgroup Just after genome duplication Outgroup More time after genome duplication Outgroup Problem (extreme case); how to solve? Outgroup Outgroup Outgroup Outgroup Outgroup Using other genomes Wong et al. 2002 PNAS Centromeres Vertebrate genome duplication Nature. 2011 Apr 10. [Epub ahead of print] Ancestral polyploidy in seed plants and angiosperms. Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, Depamphilis CW. Flowering plants Flowering MOSS Vertebrates Teleosts S. serevisiae and close relatives Paramecium Reconstructed map of genome duplications allows unprecedented mapping -

The Diversity of Plant Sex Chromosomes Highlighted Through Advances in Genome Sequencing
G C A T T A C G G C A T genes Review The Diversity of Plant Sex Chromosomes Highlighted through Advances in Genome Sequencing Sarah Carey 1,2 , Qingyi Yu 3,* and Alex Harkess 1,2,* 1 Department of Crop, Soil, and Environmental Sciences, Auburn University, Auburn, AL 36849, USA; [email protected] 2 HudsonAlpha Institute for Biotechnology, Huntsville, AL 35806, USA 3 Texas A&M AgriLife Research, Texas A&M University System, Dallas, TX 75252, USA * Correspondence: [email protected] (Q.Y.); [email protected] (A.H.) Abstract: For centuries, scientists have been intrigued by the origin of dioecy in plants, characterizing sex-specific development, uncovering cytological differences between the sexes, and developing theoretical models. Through the invention and continued improvements in genomic technologies, we have truly begun to unlock the genetic basis of dioecy in many species. Here we broadly review the advances in research on dioecy and sex chromosomes. We start by first discussing the early works that built the foundation for current studies and the advances in genome sequencing that have facilitated more-recent findings. We next discuss the analyses of sex chromosomes and sex-determination genes uncovered by genome sequencing. We synthesize these results to find some patterns are emerging, such as the role of duplications, the involvement of hormones in sex-determination, and support for the two-locus model for the origin of dioecy. Though across systems, there are also many novel insights into how sex chromosomes evolve, including different sex-determining genes and routes to suppressed recombination. We propose the future of research in plant sex chromosomes should involve interdisciplinary approaches, combining cutting-edge technologies with the classics Citation: Carey, S.; Yu, Q.; to unravel the patterns that can be found across the hundreds of independent origins. -

Cytogenetics of Fraxinus Mandshurica and F. Quadrangulata: Ploidy Determination and Rdna Analysis
Tree Genetics & Genomes (2020) 16:26 https://doi.org/10.1007/s11295-020-1418-6 ORIGINAL ARTICLE Cytogenetics of Fraxinus mandshurica and F. quadrangulata: ploidy determination and rDNA analysis Nurul Islam-Faridi1,2 & Mary E. Mason3 & Jennifer L. Koch4 & C. Dana Nelson5,6 Received: 22 July 2019 /Revised: 1 January 2020 /Accepted: 16 January 2020 # The Author(s) 2020 Abstract Ashes (Fraxinus spp.) are important hardwood tree species in rural, suburban, and urban forests of the eastern USA. Unfortunately, emerald ash borer (EAB, Agrilus planipennis) an invasive insect pest that was accidentally imported from Asia in the late 1980s–early 1990s is destroying them at an alarming rate. All North American ashes are highly susceptible to EAB, although blue ash (F. quadrangulata) may have some inherent attributes that provide it some protection. In contrast Manchurian ash (F. mandshurica) is relatively resistant to EAB having coevolved with the insect pest in its native range in Asia. Given its level of resistance, Manchurian ash has been considered for use in interspecies breeding programs designed to transfer resistance to susceptible North American ash species. One prerequisite for successful interspecies breeding is consistency in chromosome ploidy level and number between the candidate species. In the current study, we cytologically determined that both Manchurian ash and blue ash are diploids (2n) and have the same number of chromosomes (2n =2x = 46). We also characterized these species’ ribosomal gene families (45S and 5S rDNA) using fluorescence in situ hybridization (FISH). Both Manchurian and blue ash showed two 45S rDNA and one 5S rDNA sites, but blue ash appears to have an additional site of 45S rDNA. -

Genetic Variation in Polyploid Forage Grass: Assessing the Molecular Genetic Variability in the Paspalum Genus Cidade Et Al
Genetic variation in polyploid forage grass: Assessing the molecular genetic variability in the Paspalum genus Cidade et al. Cidade et al. BMC Genetics 2013, 14:50 http://www.biomedcentral.com/1471-2156/14/50 Cidade et al. BMC Genetics 2013, 14:50 http://www.biomedcentral.com/1471-2156/14/50 RESEARCH ARTICLE Open Access Genetic variation in polyploid forage grass: Assessing the molecular genetic variability in the Paspalum genus Fernanda W Cidade1, Bianca BZ Vigna2, Francisco HD de Souza2, José Francisco M Valls3, Miguel Dall’Agnol4, Maria I Zucchi5, Tatiana T de Souza-Chies6 and Anete P Souza1,7* Abstract Background: Paspalum (Poaceae) is an important genus of the tribe Paniceae, which includes several species of economic importance for foraging, turf and ornamental purposes, and has a complex taxonomical classification. Because of the widespread interest in several species of this genus, many accessions have been conserved in germplasm banks and distributed throughout various countries around the world, mainly for the purposes of cultivar development and cytogenetic studies. Correct identification of germplasms and quantification of their variability are necessary for the proper development of conservation and breeding programs. Evaluation of microsatellite markers in different species of Paspalum conserved in a germplasm bank allowed assessment of the genetic differences among them and assisted in their proper botanical classification. Results: Seventeen new polymorphic microsatellites were developed for Paspalum atratum Swallen and Paspalum notatum Flüggé, twelve of which were transferred to 35 Paspalum species and used to evaluate their variability. Variable degrees of polymorphism were observed within the species. Based on distance-based methods and a Bayesian clustering approach, the accessions were divided into three main species groups, two of which corresponded to the previously described Plicatula and Notata Paspalum groups. -

Nicotine Induces Polyspermy in Sea Urchin Eggs Through a Non-Cholinergic Pathway Modulating Actin Dynamics
cells Article Nicotine Induces Polyspermy in Sea Urchin Eggs through a Non-Cholinergic Pathway Modulating Actin Dynamics 1,2 1, 2 1, Nunzia Limatola , Filip Vasilev y, Luigia Santella and Jong Tai Chun * 1 Department of Biology and Evolution of Marine Organisms, Stazione Zoologica Anton Dohrn, I-80121 Napoli, Italy; [email protected] (N.L.); [email protected] (F.V.) 2 Department of Research Infrastructures for Marine Biological Resources, Stazione Zoologica Anton Dohrn, I-80121 Napoli, Italy; [email protected] * Correspondence: [email protected] Current address: Centre de Recherche du Centre Hospitalier de l’Université de Montreal (CRCHUM) y Montreal, QC H2X 0A9, Canada. Received: 8 November 2019; Accepted: 21 December 2019; Published: 25 December 2019 Abstract: While alkaloids often exert unique pharmacological effects on animal cells, exposure of sea urchin eggs to nicotine causes polyspermy at fertilization in a dose-dependent manner. Here, we studied molecular mechanisms underlying the phenomenon. Although nicotine is an agonist of ionotropic acetylcholine receptors, we found that nicotine-induced polyspermy was neither mimicked by acetylcholine and carbachol nor inhibited by specific antagonists of nicotinic acetylcholine receptors. Unlike acetylcholine and carbachol, nicotine uniquely induced drastic rearrangement of egg cortical microfilaments in a dose-dependent way. Such cytoskeletal changes appeared to render the eggs more receptive to sperm, as judged by the significant alleviation of polyspermy by latrunculin-A and mycalolide-B. In addition, our fluorimetric assay provided the first evidence that nicotine directly accelerates polymerization kinetics of G-actin and attenuates depolymerization of preassembled F-actin. Furthermore, nicotine inhibited cofilin-induced disassembly of F-actin. -

The Longest Telomeres: a General Signature of Adult Stem Cell Compartments
Downloaded from genesdev.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press The longest telomeres: a general signature of adult stem cell compartments Ignacio Flores,1 Andres Canela,1 Elsa Vera,1 Agueda Tejera,1 George Cotsarelis,2 and María A. Blasco1,3 1Telomeres and Telomerase Group, Molecular Oncology Program, Spanish National Cancer Centre (CNIO), Madrid E-28029, Spain; 2University of Pennsylvania School of Medicine, M8 Stellar-Chance Laboratories, Philadelphia, Pennsylvania 19104, USA Identification of adult stem cells and their location (niches) is of great relevance for regenerative medicine. However, stem cell niches are still poorly defined in most adult tissues. Here, we show that the longest telomeres are a general feature of adult stem cell compartments. Using confocal telomere quantitative fluorescence in situ hybridization (telomapping), we find gradients of telomere length within tissues, with the longest telomeres mapping to the known stem cell compartments. In mouse hair follicles, we show that cells with the longest telomeres map to the known stem cell compartments, colocalize with stem cell markers, and behave as stem cells upon treatment with mitogenic stimuli. Using K15-EGFP reporter mice, which mark hair follicle stem cells, we show that GFP-positive cells have the longest telomeres. The stem cell compartments in small intestine, testis, cornea, and brain of the mouse are also enriched in cells with the longest telomeres. This constitutes the description of a novel general property of adult stem cell compartments. Finally, we make the novel finding that telomeres shorten with age in different mouse stem cell compartments, which parallels a decline in stem cell functionality, suggesting that telomere loss may contribute to stem cell dysfunction with age. -

Arabidopsis MZT1 Homologs GIP1 and GIP2 Are Essential for Centromere Architecture
Arabidopsis MZT1 homologs GIP1 and GIP2 are essential for centromere architecture Morgane Batzenschlagera, Inna Lermontovab, Veit Schubertb, Jörg Fuchsb, Alexandre Berra, Maria A. Koinic, Guy Houlnéa, Etienne Herzoga, Twan Ruttenb, Abdelmalek Aliouaa, Paul Franszc, Anne-Catherine Schmita, and Marie-Edith Chaboutéa,1 aInstitut de biologie moléculaire des plantes, CNRS, Université de Strasbourg, 67000 Strasbourg, France; bLeibniz Institute of Plant Genetics and Crop Plant Research OT Gatersleben, D-06466 Stadt Seeland, Germany; and cSwammerdam Institute for Life Sciences, University of Amsterdam, 1098 XH, Amsterdam, The Netherlands Edited by James A. Birchler, University of Missouri, Columbia, MO, and approved May 12, 2015 (received for review April 2, 2015) Centromeres play a pivotal role in maintaining genome integrity Previously, we characterized the γ-tubulin complex protein 3- by facilitating the recruitment of kinetochore and sister-chromatid interacting proteins (GIPs), GIP1 and GIP2 (Table S1), as es- cohesion proteins, both required for correct chromosome segre- sential for the recruitment of γ-tubulin complexes at microtubule gation. Centromeres are epigenetically specified by the presence (MT) organizing centers in Arabidopsis (7, 8). This function seems of the histone H3 variant (CENH3). In this study, we investigate the conservedinthehumanandSchizosaccharomyces pombe GIP role of the highly conserved γ-tubulin complex protein 3-interact- homologs named mitotic spindle organizing protein 1 (MZT1) ing proteins (GIPs) in Arabidopsis centromere regulation. We show (9–11). More recently, we localized GIPs at the nucleoplasm pe- that GIPs form a complex with CENH3 in cycling cells. GIP depletion riphery, close to chromocenters, where they modulate the nuclear in the gip1gip2 knockdown mutant leads to a decreased CENH3 architecture (12, 13). -

Density-Dependent Sexual Selection in External Fertilizers
vol. 164, no. 3 the american naturalist september 2004 ൴ Density-Dependent Sexual Selection in External Fertilizers: Variances in Male and Female Fertilization Success along the Continuum from Sperm Limitation to Sexual Conflict in the Sea Urchin Strongylocentrotus franciscanus Don R. Levitan* Department of Biological Science, Florida State University, Keywords: sexual selection, density dependence, echinoid, micro- Tallahassee, Florida 32306-1100 satellites, multiple paternity, Bateman’s principle. Submitted October 31, 2003; Accepted May 4, 2004; Electronically published July XX, 2004 Sexual selection, the selection that arises from differences in mating success (Arnold 1994b), can strongly influence Online enhancements: appendix tables. the evolution of traits within a species and the evolution of reproductive isolation among species (Andersson 1994). Most research on sexual selection has focused on situations where sexual selection is clearly more intense for one sex abstract: Sperm competition and female choice are fundamentally than the other. Because they often invest less in each suc- driven by gender differences in investment per offspring and are cessful mating, males are generally regarded as competing often manifested as differences in variance in reproductive success: for mates, and females are seen as choosing among po- males compete and have high variance; most females are mated and tential mates (Birkhead and Møller 1998). Male compe- have low variance. In marine organisms that broadcast spawn, how- tition can result in high variance in male reproductive ever, females may encounter either sperm limitation or sperm com- success because some males are more successful than oth- petition. I measured the fertilization success of male and female Strongylocentrotus franciscanus over a range of population densities ers, and it can result in low variance in female reproductive using microsatellite markers. -

Achieving Monospermy Or Preventing Polyspermy? Open Access to Scientific and Medical Research DOI
Journal name: Research and Reports in Biology Article Designation: REVIEW Year: 2016 Volume: 7 Research and Reports in Biology Dovepress Running head verso: Dale Running head recto: Achieving monospermy or preventing polyspermy? open access to scientific and medical research DOI: http://dx.doi.org/10.2147/RRB.S84085 Open Access Full Text Article REVIEW Achieving monospermy or preventing polyspermy? Brian Dale Abstract: Images of sea urchin oocytes with hundreds of spermatozoa attached to their sur- face have fascinated scientists for over a century and led to the idea that oocytes have evolved Centre for Assisted Fertilization, Naples, Italy mechanisms to allow the penetration of one spermatozoon while repelling supernumerary spermatozoa. Popular texts have extrapolated this concept, to the mammals and amphibians, and in many cases to include all the Phyla. Here, it is argued that laboratory experiments, using sea urchin oocytes deprived of their extracellular coats and inseminated at high densities, are artifactual and that the experiments leading to the idea of a fast block to polyspermy are flawed. Under natural conditions, the number of spermatozoa at the site of fertilization is extremely low, compared with the numbers generated. The sperm:oocyte ratio is regulated first by dilution in For personal use only. externally fertilizing species and the female reproductive tract in those with internal fertiliza- tion, followed by a bottleneck created by the oocytes extracellular coats. In order to progress to the oocyte plasma membrane, the fertilizing spermatozoon must encounter and respond to a correct sequence of signals from the oocytes extracellular coats. Those that do not, are halted in their progression by defective signaling and fall to the wayside. -

Worldwide Genetic Variation in Dopamine and Serotonin Pathway
American Journal of Medical Genetics Part B (Neuropsychiatric Genetics) 147B:1070–1075 (2008) Worldwide Genetic Variation in Dopamine and Serotonin Pathway Genes: Implications for Association Studies Michelle Gardner,1,2 Jaume Bertranpetit,1,2 and David Comas1,2* 1Unitat de Biologia Evolutiva, Universitat Pompeu Fabra, Doctor Aiguader, Barcelona, Spain 2CIBER Epidemiologı´a y Salud Pu´blica (CIBERESP), Barcelona, Spain The dopamine and serotonin systems are two of the most important neurotransmitter pathways in Please cite this article as follows: Gardner M, Bertran- the human nervous system and their roles in petit J, Comas D. 2008. Worldwide Genetic Variation controlling behavior and mental status are well in Dopamine and Serotonin Pathway Genes: Implica- accepted. Genes from both systems have been tions for Association Studies. Am J Med Genet widely implicated in psychiatric and behavioral Part B 147B:1070–1075. disorders, with numerous reports of associations and almost equally as numerous reports of the failure to replicate a previous finding of associa- tion. We investigate a set of 21 dopamine and serotonin genes commonly tested for association INTRODUCTION with psychiatric disease in a set of 39 worldwide The dopamine and serotonin systems are two of the major populations representing global genetic diversity neurotransmitter systems in humans. Dopamine affects brain to see whether the failure to replicate findings of processes that control both motor and emotional behavior and association may be explained by population based it is known to have a role in the brain’s reward mechanism differences in allele frequencies and linkage [Schultz, 2002]. Serotonin has a critical role in temperature disequilibrium (LD) in this gene set.