2U12/U37116 A2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

WO 2019/068007 Al Figure 2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/068007 Al 04 April 2019 (04.04.2019) W 1P O PCT (51) International Patent Classification: (72) Inventors; and C12N 15/10 (2006.01) C07K 16/28 (2006.01) (71) Applicants: GROSS, Gideon [EVIL]; IE-1-5 Address C12N 5/10 (2006.0 1) C12Q 1/6809 (20 18.0 1) M.P. Korazim, 1292200 Moshav Almagor (IL). GIBSON, C07K 14/705 (2006.01) A61P 35/00 (2006.01) Will [US/US]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., C07K 14/725 (2006.01) P.O. Box 4044, 7403635 Ness Ziona (TL). DAHARY, Dvir [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (21) International Application Number: Box 4044, 7403635 Ness Ziona (IL). BEIMAN, Merav PCT/US2018/053583 [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (22) International Filing Date: Box 4044, 7403635 Ness Ziona (E.). 28 September 2018 (28.09.2018) (74) Agent: MACDOUGALL, Christina, A. et al; Morgan, (25) Filing Language: English Lewis & Bockius LLP, One Market, Spear Tower, SanFran- cisco, CA 94105 (US). (26) Publication Language: English (81) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of national protection available): AE, AG, AL, AM, 62/564,454 28 September 2017 (28.09.2017) US AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, 62/649,429 28 March 2018 (28.03.2018) US CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (71) Applicant: IMMP ACT-BIO LTD. -

Olfactory Receptors in Non-Chemosensory Tissues
BMB Reports Invited Mini Review Olfactory receptors in non-chemosensory tissues NaNa Kang & JaeHyung Koo* Department of Brain Science, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu 711-873, Korea Olfactory receptors (ORs) detect volatile chemicals that lead to freezing behavior (3-5). the initial perception of smell in the brain. The olfactory re- ORs are localized in the cilia of olfactory sensory neurons ceptor (OR) is the first protein that recognizes odorants in the (OSNs) in the olfactory epithelium (OE) and are activated by olfactory signal pathway and it is present in over 1,000 genes chemical cues, typically odorants at the molecular level, in mice. It is also the largest member of the G protein-coupled which lead to the perception of smell in the brain (6). receptors (GPCRs). Most ORs are extensively expressed in the Tremendous research was conducted since Buck and Axel iso- nasal olfactory epithelium where they perform the appropriate lated ORs as an OE-specific expression in 1991 (7). OR genes, physiological functions that fit their location. However, recent the largest family among the G protein-coupled receptors whole-genome sequencing shows that ORs have been found (GPCRs) (8), constitute more than 1,000 genes on the mouse outside of the olfactory system, suggesting that ORs may play chromosome (9, 10) and more than 450 genes in the human an important role in the ectopic expression of non-chemo- genome (11, 12). sensory tissues. The ectopic expressions of ORs and their phys- Odorant activation shows a distinct signal transduction iological functions have attracted more attention recently since pathway for odorant perception. -

Sean Raspet – Molecules
1. Commercial name: Fructaplex© IUPAC Name: 2-(3,3-dimethylcyclohexyl)-2,5,5-trimethyl-1,3-dioxane SMILES: CC1(C)CCCC(C1)C2(C)OCC(C)(C)CO2 Molecular weight: 240.39 g/mol Volume (cubic Angstroems): 258.88 Atoms number (non-hydrogen): 17 miLogP: 4.43 Structure: Biological Properties: Predicted Druglikenessi: GPCR ligand -0.23 Ion channel modulator -0.03 Kinase inhibitor -0.6 Nuclear receptor ligand 0.15 Protease inhibitor -0.28 Enzyme inhibitor 0.15 Commercial name: Fructaplex© IUPAC Name: 2-(3,3-dimethylcyclohexyl)-2,5,5-trimethyl-1,3-dioxane SMILES: CC1(C)CCCC(C1)C2(C)OCC(C)(C)CO2 Predicted Olfactory Receptor Activityii: OR2L13 83.715% OR1G1 82.761% OR10J5 80.569% OR2W1 78.180% OR7A2 77.696% 2. Commercial name: Sylvoxime© IUPAC Name: N-[4-(1-ethoxyethenyl)-3,3,5,5tetramethylcyclohexylidene]hydroxylamine SMILES: CCOC(=C)C1C(C)(C)CC(CC1(C)C)=NO Molecular weight: 239.36 Volume (cubic Angstroems): 252.83 Atoms number (non-hydrogen): 17 miLogP: 4.33 Structure: Biological Properties: Predicted Druglikeness: GPCR ligand -0.6 Ion channel modulator -0.41 Kinase inhibitor -0.93 Nuclear receptor ligand -0.17 Protease inhibitor -0.39 Enzyme inhibitor 0.01 Commercial name: Sylvoxime© IUPAC Name: N-[4-(1-ethoxyethenyl)-3,3,5,5tetramethylcyclohexylidene]hydroxylamine SMILES: CCOC(=C)C1C(C)(C)CC(CC1(C)C)=NO Predicted Olfactory Receptor Activity: OR52D1 71.900% OR1G1 70.394% 0R52I2 70.392% OR52I1 70.390% OR2Y1 70.378% 3. Commercial name: Hyperflor© IUPAC Name: 2-benzyl-1,3-dioxan-5-one SMILES: O=C1COC(CC2=CC=CC=C2)OC1 Molecular weight: 192.21 g/mol Volume -
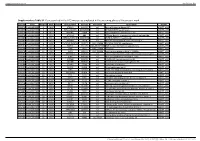
Supplementary Table S1. Genes Printed in the HC5 Microarray Employed in the Screening Phase of the Present Work
Supplementary material Ann Rheum Dis Supplementary Table S1. Genes printed in the HC5 microarray employed in the screening phase of the present work. CloneID Plate Position Well Length GeneSymbol GeneID Accession Description Vector 692672 HsxXG013989 2 B01 STK32A 202374 null serine/threonine kinase 32A pANT7_cGST 692675 HsxXG013989 3 C01 RPS10-NUDT3 100529239 null RPS10-NUDT3 readthrough pANT7_cGST 692678 HsxXG013989 4 D01 SPATA6L 55064 null spermatogenesis associated 6-like pANT7_cGST 692679 HsxXG013989 5 E01 ATP1A4 480 null ATPase, Na+/K+ transporting, alpha 4 polypeptide pANT7_cGST 692689 HsxXG013989 6 F01 ZNF816-ZNF321P 100529240 null ZNF816-ZNF321P readthrough pANT7_cGST 692691 HsxXG013989 7 G01 NKAIN1 79570 null Na+/K+ transporting ATPase interacting 1 pANT7_cGST 693155 HsxXG013989 8 H01 TNFSF12-TNFSF13 407977 NM_172089 TNFSF12-TNFSF13 readthrough pANT7_cGST 693161 HsxXG013989 9 A02 RAB12 201475 NM_001025300 RAB12, member RAS oncogene family pANT7_cGST 693169 HsxXG013989 10 B02 SYN1 6853 NM_133499 synapsin I pANT7_cGST 693176 HsxXG013989 11 C02 GJD3 125111 NM_152219 gap junction protein, delta 3, 31.9kDa pANT7_cGST 693181 HsxXG013989 12 D02 CHCHD10 400916 null coiled-coil-helix-coiled-coil-helix domain containing 10 pANT7_cGST 693184 HsxXG013989 13 E02 IDNK 414328 null idnK, gluconokinase homolog (E. coli) pANT7_cGST 693187 HsxXG013989 14 F02 LYPD6B 130576 null LY6/PLAUR domain containing 6B pANT7_cGST 693189 HsxXG013989 15 G02 C8orf86 389649 null chromosome 8 open reading frame 86 pANT7_cGST 693194 HsxXG013989 16 H02 CENPQ 55166 -

Functional Variability in the Human Odorant Receptor Repertoire
ART ic LE s The missense of smell: functional variability in the human odorant receptor repertoire Joel D Mainland1–3, Andreas Keller4, Yun R Li2,6, Ting Zhou2, Casey Trimmer1, Lindsey L Snyder1, Andrew H Moberly1,3, Kaylin A Adipietro2, Wen Ling L Liu2, Hanyi Zhuang2,6, Senmiao Zhan2, Somin S Lee2,6, Abigail Lin2 & Hiroaki Matsunami2,5 Humans have ~400 intact odorant receptors, but each individual has a unique set of genetic variations that lead to variation in olfactory perception. We used a heterologous assay to determine how often genetic polymorphisms in odorant receptors alter receptor function. We identified agonists for 18 odorant receptors and found that 63% of the odorant receptors we examined had polymorphisms that altered in vitro function. On average, two individuals have functional differences at over 30% of their odorant receptor alleles. To show that these in vitro results are relevant to olfactory perception, we verified that variations in OR10G4 genotype explain over 15% of the observed variation in perceived intensity and over 10% of the observed variation in perceived valence for the high-affinity in vitro agonist guaiacol but do not explain phenotype variation for the lower-affinity agonists vanillin and ethyl vanillin. The human genome contains ~800 odorant receptor genes that have RESULTS been shown to exhibit high genetic variability1–3. In addition, humans High-throughput screening of human odorant receptors exhibit considerable variation in the perception of odorants4,5, and To identify agonists for a variety of odorant receptors, we cloned a variation in an odorant receptor predicts perception in four cases: library of 511 human odorant receptor genes for a high-throughput loss of function in OR11H7P, OR2J3, OR5A1 and OR7D4 leads to heterologous screen. -

1 Novel Expression Signatures Identified by Transcriptional Analysis
ARD Online First, published on October 7, 2009 as 10.1136/ard.2009.108043 Ann Rheum Dis: first published as 10.1136/ard.2009.108043 on 7 October 2009. Downloaded from Novel expression signatures identified by transcriptional analysis of separated leukocyte subsets in SLE and vasculitis 1Paul A Lyons, 1Eoin F McKinney, 1Tim F Rayner, 1Alexander Hatton, 1Hayley B Woffendin, 1Maria Koukoulaki, 2Thomas C Freeman, 1David RW Jayne, 1Afzal N Chaudhry, and 1Kenneth GC Smith. 1Cambridge Institute for Medical Research and Department of Medicine, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK 2Roslin Institute, University of Edinburgh, Roslin, Midlothian, EH25 9PS, UK Correspondence should be addressed to Dr Paul Lyons or Prof Kenneth Smith, Department of Medicine, Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK. Telephone: +44 1223 762642, Fax: +44 1223 762640, E-mail: [email protected] or [email protected] Key words: Gene expression, autoimmune disease, SLE, vasculitis Word count: 2,906 The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in their licence (http://ard.bmj.com/ifora/licence.pdf). http://ard.bmj.com/ on September 29, 2021 by guest. Protected copyright. 1 Copyright Article author (or their employer) 2009. -

Human Olfactory Receptors
Human Olfactory Receptors: A journey from cell engineering for efficient in vitro functional assays to effective antagonists in human sensory assay Huysseune Sandra, Philippeau Magali, Moreau Cédric, Veithen Alex, Chatelain Pierre, Quesnel Yannick. ChemCom S.A., Route de Lennik 802, 1070 Anderlecht, Belgium. Cell lines Introduction: ChemCom's Entry ORs Literature data HEK293T HANA3 proprietary Many odorant compounds are perceived as unpleasant. They can be present in different contexts such as body-, home-, factory-, material-, or fabric-emitted 1 OR56A1 Adipietro et al., 2012 odors, so that humans are daily exposed to this olfactory pollution. In addition, odorant compounds can also taint food or beverages. Malodor and off-note 2 OR10J5 Saito et al., 2009 counteraction is a daily challenge for many different industries. 3 ORX116 - The first step of the odor perception corresponds to the interaction of olfactory receptors (ORs) with odorant molecules. Therefore, a selective inhibition of the 4 ORX069 - ORs by weakly odorant or odorless antagonists represents an innovative solution to malodor issues. The identification of such odor blockers requires an 5 OR10H5 - efficient technological platform to first fish out the receptors that interact with a malodor of interest and second, to screen libraries of potential antagonists of 6 OR8B3 Mainland et al., 2013 7 OR6P1 Mainland et al., 2013 these ORs. 8 ORX074B - 9 ORX126 - Materials and Methods: 10 ORX189 - 11 ORX081 - In vitro functional assay 12 ORX213 - Dilution-response analysis were performed in HEK293T, HANA3 and HEK293T-hRTP1S/hRTP2 cells using the CRE-luciferase reporter assay system. Briefly, each 13 OR1D2 Spehr et al., 2003 cell plated one day before was transfected with deorphanized ORs (identified at Chemcom S.A.) or empty vector plasmids using TransIT®-LT1 (Mirus) 14 OR7C1 Mainland et al., 2013 according to the manufacturer’s protocol. -

Table S4. RAE Analysis of Dedifferentiated Liposarcoma
Table S4. RAE analysis of dedifferentiated liposarcoma Model Chromosome Region start Region end Size q value freqX0* # genes genes Amp 1 57809872 60413476 2603605 0.00026 34.6 10 DAB1,RPS26P15,OMA1,TACSTD2,MYSM1,JUN,FGGY,HOOK1,CYP2J2,C1orf87 Amp 1 158619146 158696968 77823 0.053 25 1 VANGL2 Amp 1 158883523 158922841 39319 0.081 23.1 2 SLAMF1,CD48 Amp 1 162042586 162118557 75972 0.072 25 0 [Nearest:NUF2] Amp 1 162272460 162767627 495168 0.017 26.9 0 [Nearest:PBX1] Amp 1 165486554 165532374 45821 0.057 25 1 POU2F1 Amp 1 167138282 167483267 344986 0.024 26.9 2 ATP1B1,NME7 Amp 1 167612872 167708844 95973 0.041 25 3 BLZF1,C1orf114,SLC19A2 Amp 1 167728199 167808161 79963 0.076 21.2 1 F5 Amp 1 168436370 169233893 797524 0.018 26.9 3 GORAB,PRRX1,C1orf129 Amp 1 169462231 170768440 1306210 1.3E-06 38.5 10 FMO1,FMO4,TOP1P1,BAT2D1,MYOC,VAMP4,METTL13,DNM3,C1orf105,PIGC Amp 1 171026247 171291427 265181 0.015 26.9 1 TNFSF18 Del 1 201860394 202299299 438906 0.0047 25 6 ATP2B4,SNORA77,LAX1,ZC3H11A,SNRPE,C1orf157 Del 1 210909187 212021116 1111930 0.017 19.2 8 BATF3,NSL1,TATDN3,C1orf227,FLVCR1,VASH2,ANGEL2,RPS6KC1 Del 1 215937857 216049214 111358 0.079 23.1 1 SPATA17 Del 1 218237257 218367476 130220 0.0063 26.9 3 EPRS,BPNT1,IARS2 Del 1 222100886 222727238 626353 5.2E-05 32.7 5 FBXO28,DEGS1,NVL,CNIH4,WDR26 Del 1 223166548 224519805 1353258 0.0063 26.9 15 DNAH14,LBR,ENAH,SRP9,EPHX1,TMEM63A,LEFTY1,PYCR2,LEFTY2,C1orf55,H3F3A,LOC440926 ,ACBD3,MIXL1,LIN9 Del 1 225283136 225374166 91031 0.054 23.1 1 CDC42BPA Del 1 227278990 229012661 1733672 0.091 21.2 13 RAB4A,SPHAR,C1orf96,ACTA1,NUP133,ABCB10,TAF5L,URB2,GALNT2,PGBD5,COG2,AGT,CAP -

The Hypothalamus As a Hub for SARS-Cov-2 Brain Infection and Pathogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis Sreekala Nampoothiri1,2#, Florent Sauve1,2#, Gaëtan Ternier1,2ƒ, Daniela Fernandois1,2 ƒ, Caio Coelho1,2, Monica ImBernon1,2, Eleonora Deligia1,2, Romain PerBet1, Vincent Florent1,2,3, Marc Baroncini1,2, Florence Pasquier1,4, François Trottein5, Claude-Alain Maurage1,2, Virginie Mattot1,2‡, Paolo GiacoBini1,2‡, S. Rasika1,2‡*, Vincent Prevot1,2‡* 1 Univ. Lille, Inserm, CHU Lille, Lille Neuroscience & Cognition, DistAlz, UMR-S 1172, Lille, France 2 LaBoratorY of Development and PlasticitY of the Neuroendocrine Brain, FHU 1000 daYs for health, EGID, School of Medicine, Lille, France 3 Nutrition, Arras General Hospital, Arras, France 4 Centre mémoire ressources et recherche, CHU Lille, LiCEND, Lille, France 5 Univ. Lille, CNRS, INSERM, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for Infection and ImmunitY of Lille (CIIL), Lille, France. # and ƒ These authors contriButed equallY to this work. ‡ These authors directed this work *Correspondence to: [email protected] and [email protected] Short title: Covid-19: the hypothalamic hypothesis 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Supplemental Table 1 (S1). the Chromosomal Regions and Genes Exhibiting Loss of Heterozygosity That Were Shared Between the HLRCC-Rccs of Both Patients 1 and 2
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Clin Pathol Supplemental Table 1 (S1). The chromosomal regions and genes exhibiting loss of heterozygosity that were shared between the HLRCC-RCCs of both Patients 1 and 2. Chromosome Position Genes 1 p13.1 ATP1A1, ATP1A1-AS1, LOC101929023, CD58, IGSF3, MIR320B1, C1orf137, CD2, PTGFRN, CD101, LOC101929099 1 p21.1* COL11A1, LOC101928436, RNPC3, AMY2B, ACTG1P4, AMY2A, AMY1A, AMY1C, AMY1B 1 p36.11 LOC101928728, ARID1A, PIGV, ZDHHC18, SFN, GPN2, GPATCH3, NR0B2, NUDC, KDF1, TRNP1, FAM46B, SLC9A1, WDTC1, TMEM222, ACTG1P20, SYTL1, MAP3K6, FCN3, CD164L2, GPR3, WASF2, AHDC1, FGR, IFI6 1 q21.3* KCNN3, PMVK, PBXIP1, PYGO2, LOC101928120, SHC1, CKS1B, MIR4258, FLAD1, LENEP, ZBTB7B, DCST2, DCST1, LOC100505666, ADAM15, EFNA4, EFNA3, EFNA1, SLC50A1, DPM3, KRTCAP2, TRIM46, MUC1, MIR92B, THBS3, MTX1, GBAP1, GBA, FAM189B, SCAMP3, CLK2, HCN3, PKLR, FDPS, RUSC1-AS1, RUSC1, ASH1L, MIR555, POU5F1P4, ASH1L-AS1, MSTO1, MSTO2P, YY1AP1, SCARNA26A, DAP3, GON4L, SCARNA26B, SYT11, RIT1, KIAA0907, SNORA80E, SCARNA4, RXFP4, ARHGEF2, MIR6738, SSR2, UBQLN4, LAMTOR2, RAB25, MEX3A, LMNA, SEMA4A, SLC25A44, PMF1, PMF1-BGLAP 1 q24.2–44* LOC101928650, GORAB, PRRX1, MROH9, FMO3, MIR1295A, MIR1295B, FMO6P, FMO2, FMO1, FMO4, TOP1P1, PRRC2C, MYOC, VAMP4, METTL13, DNM3, DNM3-IT1, DNM3OS, MIR214, MIR3120, MIR199A2, C1orf105, PIGC, SUCO, FASLG, TNFSF18, TNFSF4, LOC100506023, LOC101928673, -

Apoptotic Cells Inflammasome Activity During the Uptake of Macrophage
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology published online 20 April 2012 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2012/04/20/jimmun ol.1103760 Complement Protein C1q Directs Macrophage Polarization and Limits Inflammasome Activity during the Uptake of Apoptotic Cells Marie E. Benoit, Elizabeth V. Clarke, Pedro Morgado, Deborah A. Fraser and Andrea J. Tenner J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://www.jimmunol.org/content/suppl/2012/04/20/jimmunol.110376 0.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. Published April 20, 2012, doi:10.4049/jimmunol.1103760 The Journal of Immunology Complement Protein C1q Directs Macrophage Polarization and Limits Inflammasome Activity during the Uptake of Apoptotic Cells Marie E. Benoit, Elizabeth V. Clarke, Pedro Morgado, Deborah A. Fraser, and Andrea J. Tenner Deficiency in C1q, the recognition component of the classical complement cascade and a pattern recognition receptor involved in apoptotic cell clearance, leads to lupus-like autoimmune diseases characterized by auto-antibodies to self proteins and aberrant innate immune cell activation likely due to impaired clearance of apoptotic cells. -

Deciphering the Wisent Demographic and Adaptive Histories From
Deciphering the Wisent Demographic and Adaptive Histories from Individual Whole-Genome Sequences Mathieu Gautier,1,2 Katayoun Moazami-Goudarzi,3 Hubert Leve´ziel,4 Hugues Parinello,5 Ce´cile Grohs,3 Ste´phanie Rialle,5 Rafał Kowalczyk,6 and Laurence Flori*,3,7 1CBGP, INRA, CIRAD, IRD, Supagro, Montferrier-sur-Lez, France 2IBC, Institut de Biologie Computationnelle, Montpellier, France 3GABI, INRA, AgroParisTech, Universite´ Paris-Saclay, Jouy-en-Josas, France 4UGMA, INRA, Universite´ de Limoges, Limoges, France 5MGX-Montpellier GenomiX, c/o Institut de Ge´nomique Fonctionnelle, Montpellier, France 6Mammal Research Institute, Polish Academy of Sciences, Białowieza,_ Poland 7INTERTRYP, CIRAD, IRD, Montpellier, France *Corresponding author: Email: laurence.fl[email protected]. Associate editor: Emma Teeling Sequencing data (bam files) are available for download from the Sequence Read Archive repository (http://www.ncbi.nlm.nih.gov/sra) (accession no. SRP070526). Abstract As the largest European herbivore, the wisent (Bison bonasus) is emblematic of the continent wildlife but has unclear origins. Here, we infer its demographic and adaptive histories from two individual whole-genome sequences via a detailed comparativeanalysiswithbovinegenomes.Weestimatethatthewisentandbovinespeciesdivergedfrom1.7Â 106 to 850,000 years before present (YBP) through a speciation process involving an extended period of limited gene flow. Our data further support the occurrence of more recent secondary contacts, posterior to the Bos taurus and Bos indicus divergence (150,000 YBP), between the wisent and (European) taurine cattle lineages. Although the wisent and bovine population sizes experienced a similar sharp decline since the Last Glacial Maximum, we find that the wisent demography remained more fluctuating during the Pleistocene. This is in agreement with a scenario in which wisents responded to successive glaciations by habitat fragmentation rather than southward and eastward migration as for the bovine ances- tors.