Molecular Recognition Between Ketosynthase and Acyl Carrier Protein Domains of the 6-Deoxyerythronolide B Synthase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Contig Protein Description Symbol Anterior Posterior Ratio
Table S2. List of proteins detected in anterior and posterior intestine pooled samples. Data on protein expression are mean ± SEM of 4 pools fed the experimental diets. The number of the contig in the Sea Bream Database (http://nutrigroup-iats.org/seabreamdb) is indicated. Contig Protein Description Symbol Anterior Posterior Ratio Ant/Pos C2_6629 1,4-alpha-glucan-branching enzyme GBE1 0.88±0.1 0.91±0.03 0.98 C2_4764 116 kDa U5 small nuclear ribonucleoprotein component EFTUD2 0.74±0.09 0.71±0.05 1.03 C2_299 14-3-3 protein beta/alpha-1 YWHAB 1.45±0.23 2.18±0.09 0.67 C2_268 14-3-3 protein epsilon YWHAE 1.28±0.2 2.01±0.13 0.63 C2_2474 14-3-3 protein gamma-1 YWHAG 1.8±0.41 2.72±0.09 0.66 C2_1017 14-3-3 protein zeta YWHAZ 1.33±0.14 4.41±0.38 0.30 C2_34474 14-3-3-like protein 2 YWHAQ 1.3±0.11 1.85±0.13 0.70 C2_4902 17-beta-hydroxysteroid dehydrogenase 14 HSD17B14 0.93±0.05 2.33±0.09 0.40 C2_3100 1-acylglycerol-3-phosphate O-acyltransferase ABHD5 ABHD5 0.85±0.07 0.78±0.13 1.10 C2_15440 1-phosphatidylinositol phosphodiesterase PLCD1 0.65±0.12 0.4±0.06 1.65 C2_12986 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 PLCD1 0.76±0.08 1.15±0.16 0.66 C2_4412 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 PLCG2 1.13±0.08 2.08±0.27 0.54 C2_3170 2,4-dienoyl-CoA reductase, mitochondrial DECR1 1.16±0.1 0.83±0.03 1.39 C2_1520 26S protease regulatory subunit 10B PSMC6 1.37±0.21 1.43±0.04 0.96 C2_4264 26S protease regulatory subunit 4 PSMC1 1.2±0.2 1.78±0.08 0.68 C2_1666 26S protease regulatory subunit 6A PSMC3 1.44±0.24 1.61±0.08 -
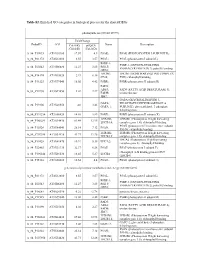
Table S2. Enriched GO Categories in Biological Process for the Shared Degs
Table S2. Enriched GO categories in biological process for the shared DEGs photosynthesis (GO ID:15979) Fold Change ProbeID AGI Col-0(R) pifQ(D) Name Description /Col-0(D) /Col-0(D) A_84_P19035 AT1G30380 17.07 4.9 PSAK; PSAK (PHOTOSYSTEM I SUBUNIT K) A_84_P21372 AT4G12800 8.55 3.57 PSAL; PSAL (photosystem I subunit L) PSBP-1; PSBP-1 (OXYGEN-EVOLVING A_84_P20343 AT1G06680 12.27 3.85 PSII-P; ENHANCER PROTEIN 2); poly(U) binding OEE2; LHCB6; LHCB6 (LIGHT HARVESTING COMPLEX A_84_P14174 AT1G15820 23.9 6.16 CP24; PSII); chlorophyll binding A_84_P11525 AT1G79040 16.02 4.42 PSBR; PSBR (photosystem II subunit R) FAD5; ADS3; FAD5 (FATTY ACID DESATURASE 5); A_84_P19290 AT3G15850 4.02 2.27 FADB; oxidoreductase JB67; GAPA (GLYCERALDEHYDE 3- GAPA; PHOSPHATE DEHYDROGENASE A A_84_P19306 AT3G26650 4.6 3.43 GAPA-1; SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase A_84_P193234 AT2G06520 14.01 3.89 PSBX; PSBX (photosystem II subunit X) LHB1B1; LHB1B1 (Photosystem II light harvesting A_84_P160283 AT2G34430 89.44 32.95 LHCB1.4; complex gene 1.4); chlorophyll binding PSAN (photosystem I reaction center subunit A_84_P10324 AT5G64040 26.14 7.12 PSAN; PSI-N); calmodulin binding LHB1B2; LHB1B2 (Photosystem II light harvesting A_84_P207958 AT2G34420 41.71 12.26 LHCB1.5; complex gene 1.5); chlorophyll binding LHCA2 (Photosystem I light harvesting A_84_P19428 AT3G61470 10.91 5.36 LHCA2; complex gene 2); chlorophyll binding A_84_P22465 AT1G31330 32.37 6.58 PSAF; PSAF (photosystem I subunit F) chlorophyll A-B binding protein CP29 A_84_P190244 AT5G01530 16.45 5.27 LHCB4 -

Fatty Acid Biosynthesis
BI/CH 422/622 ANABOLISM OUTLINE: Photosynthesis Carbon Assimilation – Calvin Cycle Carbohydrate Biosynthesis in Animals Gluconeogenesis Glycogen Synthesis Pentose-Phosphate Pathway Regulation of Carbohydrate Metabolism Anaplerotic reactions Biosynthesis of Fatty Acids and Lipids Fatty Acids contrasts Diversification of fatty acids location & transport Eicosanoids Synthesis Prostaglandins and Thromboxane acetyl-CoA carboxylase Triacylglycerides fatty acid synthase ACP priming Membrane lipids 4 steps Glycerophospholipids Control of fatty acid metabolism Sphingolipids Isoprene lipids: Cholesterol ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1 ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1. Biosynthesis of fatty acids 2. Regulation of fatty acid degradation and synthesis 3. Assembly of fatty acids into triacylglycerol and phospholipids 4. Metabolism of isoprenes a. Ketone bodies and Isoprene biosynthesis b. Isoprene polymerization i. Cholesterol ii. Steroids & other molecules iii. Regulation iv. Role of cholesterol in human disease ANABOLISM II: Biosynthesis of Fatty Acids & Lipids Lipid Fat Biosynthesis Catabolism Fatty Acid Fatty Acid Degradation Synthesis Ketone body Isoprene Utilization Biosynthesis 2 Catabolism Fatty Acid Biosynthesis Anabolism • Contrast with Sugars – Lipids have have hydro-carbons not carbo-hydrates – more reduced=more energy – Long-term storage vs short-term storage – Lipids are essential for structure in ALL organisms: membrane phospholipids • Catabolism of fatty acids –produces acetyl-CoA –produces reducing -

Saturated Long-Chain Fatty Acid-Producing Bacteria Contribute
Zhao et al. Microbiome (2018) 6:107 https://doi.org/10.1186/s40168-018-0492-6 RESEARCH Open Access Saturated long-chain fatty acid-producing bacteria contribute to enhanced colonic motility in rats Ling Zhao1†, Yufen Huang2†, Lin Lu1†, Wei Yang1, Tao Huang1, Zesi Lin3, Chengyuan Lin1,4, Hiuyee Kwan1, Hoi Leong Xavier Wong1, Yang Chen5, Silong Sun2, Xuefeng Xie2, Xiaodong Fang2,5, Huanming Yang6, Jian Wang6, Lixin Zhu7* and Zhaoxiang Bian1* Abstract Background: The gut microbiota is closely associated with gastrointestinal (GI) motility disorder, but the mechanism(s) by which bacteria interact with and affect host GI motility remains unclear. In this study, through using metabolomic and metagenomic analyses, an animal model of neonatal maternal separation (NMS) characterized by accelerated colonic motility and gut dysbiosis was used to investigate the mechanism underlying microbiota-driven motility dysfunction. Results: An excess of intracolonic saturated long-chain fatty acids (SLCFAs) was associated with enhanced bowel motility in NMS rats. Heptadecanoic acid (C17:0) and stearic acid (C18:0), as the most abundant odd- and even- numbered carbon SLCFAs in the colon lumen, can promote rat colonic muscle contraction and increase stool frequency. Increase of SLCFAs was positively correlated with elevated abundances of Prevotella, Lactobacillus, and Alistipes. Functional annotation found that the level of bacterial LCFA biosynthesis was highly enriched in NMS group. Essential synthetic genes Fabs were largely identified from the genera Prevotella, Lactobacillus, and Alistipes. Pseudo germ-free (GF) rats receiving fecal microbiota from NMS donors exhibited increased defecation frequency and upregulated bacterial production of intracolonic SLCFAs. Modulation of gut dysbiosis by neomycin effectively attenuated GI motility and reduced bacterial SLCFA generation in the colon lumen of NMS rats. -

Crystal Structure of Fabz-ACP Complex Reveals a Dynamic Seesaw-Like Catalytic Mechanism of Dehydratase in Fatty Acid Biosynthesis
Cell Research (2016) 26:1330-1344. ORIGINAL ARTICLE www.nature.com/cr Crystal structure of FabZ-ACP complex reveals a dynamic seesaw-like catalytic mechanism of dehydratase in fatty acid biosynthesis Lin Zhang1, 2, Jianfeng Xiao3, Jianrong Xu1, Tianran Fu1, Zhiwei Cao1, Liang Zhu1, 2, Hong-Zhuan Chen1, 2, Xu Shen3, Hualiang Jiang3, Liang Zhang1, 2 1Department of Pharmacology and Chemical Biology, Shanghai Jiao Tong University School of Medicine, Shanghai, China; 2Shanghai Universities Collaborative Innovation Center for Translational Medicine, Shanghai, China; 3State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China Fatty acid biosynthesis (FAS) is a vital process in cells. Fatty acids are essential for cell assembly and cellular me- tabolism. Abnormal FAS directly correlates with cell growth delay and human diseases, such as metabolic syndromes and various cancers. The FAS system utilizes an acyl carrier protein (ACP) as a transporter to stabilize and shuttle the growing fatty acid chain throughout enzymatic modules for stepwise catalysis. Studying the interactions between enzymatic modules and ACP is, therefore, critical for understanding the biological function of the FAS system. How- ever, the information remains unclear due to the high flexibility of ACP and its weak interaction with enzymatic mod- ules. We present here a 2.55 Å crystal structure of type II FAS dehydratase FabZ in complex with holo-ACP, which exhibits a highly symmetrical FabZ hexamer-ACP3 stoichiometry with each ACP binding to a FabZ dimer subunit. Further structural analysis, together with biophysical and computational results, reveals a novel dynamic seesaw-like ACP binding and catalysis mechanism for the dehydratase module in the FAS system, which is regulated by a critical gatekeeper residue (Tyr100 in FabZ) that manipulates the movements of the β-sheet layer. -

Data-Driven Insights Into Ligands, Proteins, and Genetic Mutations
Data-Driven Insights into Ligands, Proteins, and Genetic Mutations by Jing Lu A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Bioinformatics) in the University of Michigan 2016 Doctoral Committee: Professor Heather A. Carlson, Chair Professor Charles L. Brooks III Assistant Professor Barry Grant Professor David S. Sept Professor Kerby A. Shedden © Jing Lu, 2016 Acknowledgements I would like to thank my advisor, Dr. Heather Carlson, for years of patient guidance, teaching, and support through the course of my PhD. I have learnt how to think critically and be rigorous in every step of research. I also want to express gratitude to my committee: Professor Charles L. Brooks III, Assistant Professor Barry Grant, Professor David S. Sept, Professor Kerby A. Shedden. Their advising is insightful and deepens my understanding of my research projects. I would like to thank Dr. Richard Smith for timely support for both my writing and research. For many Saturdays and Sundays, he promptly responds my requests for proofreading. Much of my work is built on his code in protein and ligand analysis. I would like to thank other members in Dr. Carlson’s lab for helping me with my work. Through the discussion with Dr. Jim Dunbar, I have learnt many critical ideas in Cheminformatics. Also, thank you to Sarah Graham and Jordan Clark for their tremendous friendship and willing to help with my writing. I would also thank previous members in Dr. Carlson’s lab. I would thank Dr. Phani Ghanakota for many late-night discussions and Dr. -

Thromboplastin, Tissue Factor
TNF_repressed genes, fat tissue, 1 day infusion Description Accession Fold Blood Coagulation Coagulation factor III (thromboplastin, tissue factor) U07619 1.4 factor XIIIa Y12502 1.5 Cell Adhesion integrin alpha chain, H36-alpha7 X65036 1.6 integrin alpha-1 X52140 1.4 Cell Cycle Control-Cell Stress growth arrest and DNA-damage-inducible (GADD45) L32591 1.5 Cell Signaling adipocyte hormone-sensitive cyclic AMP phosphodiesterase Z22867 1.5 Fibroblast growth factor receptor 1 beta-isoform S54008 1.4 Insulin-like growth factor binding protein 6 M69055 1.9 LIM domain kinase 1 isoform 1 (LIMK-1) D31873 1.4 protein kinase PASK D88190 2.2 protein phosphatase 1 beta S78218 1.4 protein phosphatase inhibitor-1 protein J05592 1.9 S-100 beta subunit S53527 1.6 Cell Stress Glutathione S-transferase 1 (theta) D10026 1.6 heat shock related protein AI171166 1.5 selenoprotein P AI230247 1.4 Cell Structure and Cytoskeleton myosin regulatory light chain isoform C S77900 1.7 Cytokine cardiotrophin-1 D78591 1.5 Transforming growth factor, beta 3 U03491 1.8 DNA Synthesis nuclear factor I/A (NFI-A1) X84210 1.4 nuclear factor I/B (NF1-B2) AB012231 1.6 nuclear factor I/B (NF1-B3) AI176488 1.6 Immune Response mast cell protease 1 precursor (RMCP-1) U67915 1.5 MHC class II A-beta RT1.B-b-beta gene M36151 2.2 MHC class II antigen RT1.B-1 beta-chain X56596 1.8 MHC class II RT1.B-alpha chain gene X07551 2.2 MHC class II RT1.u-D-alpha chain mRNA M15562 2 MHC class II-associated invariant chain X14254 2.2 MHC RT1-B region class II (Ia antigen) K02815 1.8 MHC-associated -

Advance View Proofs
J-STAGE Advance Publication Date March 25, 2021 The Journal of Poultry Science doi: 10.2141/jpsa.0200095 Fasting and Refeeding Affect the Goose Liver Transcriptome Mainly Through the PPAR Signaling Pathway Zhenzhen Chen1, Ya Xing1, Xue Fan1, Tongjun Liu1, Minmeng Zhao1, Long Liu1, Xuming Hu1, Hengmi Cui1, Tuoyu Geng1,2 and Daoqing Gong1,2 1College of Animal Science and Technology, Yangzhou University, Yangzhou 225009, Jiangsu Province, P. R. China 2Joint International Research Laboratory of Agriculture and Agri-Product Safety of the Ministry of Education of China, Yangzhou University, Yangzhou 225009, Jiangsu Province, P. R. China Running title: Nutritional status affects lipid metabolism Correspondence: Tuoyu Geng and Daoqing Gong College of Animal Science and Technology, Yangzhou University, 48 Wenhui East Road,Advance Yangzhou, Jiangsu Province 225009, View P. R. China Proofs Email: [email protected] (Tuoyu Geng); [email protected] (Daoqing Gong) J-STAGE Advance Publication Date March 25, 2021 The Journal of Poultry Science doi: 10.2141/jpsa.0200095 Abstract Nutrition and energy are essential for poultry growth and production performance. Fasting and refeeding have been widely used to study the effects of nutrition, energy, and related mechanisms in chicken. Previous studies have shown that geese have a strong capacity for fat synthesis and storage; thus, changes in the goose liver transcriptome may be different from those in chicken assessed with a model of fasting and refeeding. However, the responses of the goose liver transcriptome to fasting and refeeding have not yet been addressed. In this study, 36 70-day-old Si Ji geese with similar body weight were randomly assigned to three groups: control (ad libitum feeding), fasting (fasted for 24 h), and refeeding (fast for 24 h followed by 2-h feeding) groups. -

S1 of S77 Supplementary Materials: Discovery of a New Class of Cathepsin Kinhibitors in Rhizoma Drynariaeas Potential Candidates for the Treatment of Osteoporosis
Int. J. Mol. Sci.2016, 17, 2116; doi:10.3390/ijms17122116 S1 of S77 Supplementary Materials: Discovery of a New Class of Cathepsin KInhibitors in Rhizoma Drynariaeas Potential Candidates for the Treatment of Osteoporosis Zuo-Cheng Qiu, Xiao-Li Dong, Yi Dai, Gao-Keng Xiao, Xin-Luan Wang, Ka-Chun Wong, Man-Sau Wong and Xin-Sheng Yao Table S1. Compounds identified from Drynariae rhizome (DR). No. Compound Name Chemical Structure 1 Naringin 5,7,3′,5′-Tetrahydroxy-flavanone 2 7-O-neohesperidoside 3 Narigenin-7-O-β-D-glucoside 5,7,3′,5′-Tetrahydroxy-flavanone 4 7-O-β-D-glucopyranoside 5 Naringenin 6 5,7,3′,5′-Tetrahydroxyflavanone 7 Kushennol F 8 Sophoraflavanone G 9 Kurarinone Int. J. Mol. Sci.2016, 17, 2116; doi:10.3390/ijms17122116 S2 of S77 Table S1. Cont. No. Compound Name Chemical Structure 10 Leachianone A 11 Luteolin-7-O-neohesperidoside 12 Luteolin-5-O-neohesperidoside 13 Kaempferol-7-O-α-L-arabinofuranoside 14 8-Prenylapigenin 15 Apigenine 16 Kaempferol-3-O-α-L-rhamnopyranoside OH HO O 17 Astragalin O OH OH O O OH OH OH 18 3-O-β-D-Glucopyranoside-7-O-α-L-arabinofuranoside OH HO O HO O O 19 5,7-Dihydroxychromone-7-O-β-D-glucopyranoside OH OH O Int. J. Mol. Sci.2016, 17, 2116; doi:10.3390/ijms17122116 S3 of S77 Table S1. Cont. No. Compound Name Chemical Structure 20 5,7-Dihydroxychromone-7-O-neohesperidoside Kaempferol 21 3-O-β-D-glucopyranoside-7-O-β-D-glucopyranoside 22 Xanthohumol OH HO O 23 Epicatechin OH OH OH 24 (E)-4-O-β-D-Glucopyranosyl caffeic acid 25 β-D-Glucopyranosyl sinapoic acid 26 4-O-β-D-Glucopyranosyl ferulic acid 27 Trans-caffeic acid 28 4-O-β-D-Glucopyranosyl coumaric acid 29 Dihydrocaffeic acid methyl ester 30 Dihydrocaffeic acid 31 3,4-Dihydroxyl benzoic acid 32 4-O-D-Glucosyl vanillic acid Int. -

Deciphering the Binding Interactions Between Acinetobacter Baumannii ACP and Β-Ketoacyl ACP Synthase III to Improve Antibiotic Targeting Using NMR Spectroscopy
International Journal of Molecular Sciences Article Deciphering the Binding Interactions between Acinetobacter baumannii ACP and β-ketoacyl ACP Synthase III to Improve Antibiotic Targeting Using NMR Spectroscopy Sungjae Choi, Jungwoo Park, Jiwon Yeon, Ahjin Jang, Woo Cheol Lee and Yangmee Kim * Department of Bioscience and Biotechnology, Konkuk University, Seoul 05029, Korea; [email protected] (S.C.); [email protected] (J.P.); [email protected] (J.Y.); [email protected] (A.J.); [email protected] (W.C.L.) * Correspondence: [email protected]; Tel.: +822-450-3421; Fax: +822-447-5987 Abstract: Fatty acid synthesis is essential for bacterial viability. Thus, fatty acid synthases (FASs) represent effective targets for antibiotics. Nevertheless, multidrug-resistant bacteria, including the human opportunistic bacteria, Acinetobacter baumannii, are emerging threats. Meanwhile, the FAS pathway of A. baumannii is relatively unexplored. Considering that acyl carrier protein (ACP) has an important role in the delivery of fatty acyl intermediates to other FAS enzymes, we elucidated the solution structure of A. baumannii ACP (AbACP) and, using NMR spectroscopy, investigated its interactions with β-ketoacyl ACP synthase III (AbKAS III), which initiates fatty acid elongation. The results show that AbACP comprises four helices, while Ca2+ reduces the electrostatic repulsion between acid residues, and the unconserved F47 plays a key role in thermal stability. Moreover, AbACP exhibits flexibility near the hydrophobic cavity entrance from D59 to T65, as well as in the Citation: Choi, S.; Park, J.; Yeon, J.; α1α2 loop region. Further, F29 and A69 participate in slow exchanges, which may be related to Jang, A.; Lee, W.C.; Kim, Y. -

UC San Francisco Electronic Theses and Dissertations
UCSF UC San Francisco Electronic Theses and Dissertations Title Toward drugging the translocon: sequence determinants and cellular consequences Sec61 inhibition Permalink https://escholarship.org/uc/item/9610t3s7 Author Maglathlin, Rebecca Publication Date 2014 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California Copyright (2014) By Rebecca L. Maglathlin ii Acknowledgements “In the discovery of secret things, and in the investigation of hidden causes, stronger reasons are obtained from sure experiments and demonstrated arguments than from probable conjectures and the opinions of philosophical speculators.” -William Gilbert, Loadstone and Magnetic Bodies, and on The Great Magnet of the Earth, translated from the 1600 edition of De Magnete by P. Fleury Mottelay (Bernard Quaritch, London, 1893) I would like to thank my mentor, Jack Taunton, for instilling in me that “good” is never enough and that greatness is just as much a matter of hard work and perseverance as it is a function of intelligence and insight. I would like to thank Jeff Johnson and Tasha Johnson for their work on the mass spectrometry in Chapter 2. I would also like to thank Gonzalo Ureta and Emma McCullagh of Sebastian Bernales’ Lab (Fundacion Ciencia de la Vida, Chile) for their work cited in Chapter 3. I would like to thank the members of the Taunton Lab, past and present, for their insights, expertise, friendship and general all around awesomeness. I would specifically like to thank Ville, Sarah and Andy for their guidance on this project and for their beautiful work cited herein. I would also like to thank Geoff Smith for his contribution of the STAT5 phosphorylation experiment in Chapter 2. -

Identification and Functional Characterization of Adipogenesis-Related Genes
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Sciences Identification and Functional Characterization of Adipogenesis-related Genes Submitted by: Yu Wu In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Sciences Examination Committee Signature/Date Major Advisor: Cynthia M. Smas, D.Sc. Academic Kam Yeung, Ph.D. Advisory Committee: Ronald L. Mellgren, Ph.D. William T. Gunning, Ph.D. Beata Lecka-Czernik, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: October 14, 2008 Identification and Characterization of Adipogenesis Related Genes Yu Wu University of Toledo Health Science Campus 2008 i DEDICATION This work is dedicated to my father, Zixing Wu, and my mother, Zhiping Yan, for their great love, and to my husband Yang Lu, for his continual support, encouragement and love. ii ACKNOWLEDGEMENTS I want to express my most deep gratitude to my advisor, Dr. Cynthia M. Smas for Her guidance, understanding, patience, and encouragement in the past four years. Her infectious enthusiasm and unlimited zeal have always been the major driving forces during my graduate studies. She encouraged me to develop independent thinking and research skills, which prepared me for future challenges. I would like to extend my thanks to the distinguished faculty members who served on my committee: Dr. Kam C. Yeung, Dr. Ronald L. Mellgren, Dr. William T. Gunning III, Dr. Beata Lecka-Czernik and Dr. Ana Maria Oyarce. I have benefited greatly from their advices. This dissertation would not be realized without the help of my colleagues and friends at the University of Toledo.