UC San Francisco Electronic Theses and Dissertations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genome-Wide Association Studies Identify Genetic Loci Associated With
SUPPLEMENTARY DATA Genome-wide Association Studies Identify Genetic Loci Associated with Albuminuria in Diabetes SUPPLEMENTAL MATERIALS This work is dedicated to the memory of our colleague Dr. Wen Hong Linda Kao, a wonderful person, brilliant scientist and central member of the CKDGen Consortium. 1 ©2016 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-1313/-/DC1 SUPPLEMENTARY DATA Table of Contents SUPPLEMENTARY FIGURE 1: QQ PLOTS FOR ALL GWAS META-ANALYSES ............................................. 3 SUPPLEMENTARY FIGURE 2: MANHATTAN PLOTS FOR ALL GWAS META-ANALYSES ............................. 4 SUPPLEMENTARY FIGURE 3: REGIONAL ASSOCIATION PLOTS............................................................... 6 SUPPLEMENTARY FIGURE 4: EVALUATION OF GLOMERULOSCLEROSIS IN RAB38 KO, CONGENIC AND TRANSGENIC RATS. ........................................................................................................................... 17 SUPPLEMENTARY TABLE 1: CHARACTERISTICS OF THE STUDY POPULATIONS ..................................... 18 SUPPLEMENTARY TABLE 2: INFORMATION ABOUT STUDY DESIGN AND UACR MEASUREMENT .......... 20 SUPPLEMENTARY TABLE 3: STUDY-SPECIFIC INFORMATION ABOUT GENOTYPING, IMPUTATION AND DATA MANAGEMENT AND ANALYSIS ................................................................................................ 31 SUPPLEMENTARY TABLE 4: SNPS ASSOCIATED WITH UACR AMONG ALL INDIVIDUALS WITH A P-VALUE OF <1E-05. ....................................................................................................................................... -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

1 ICR-Geneset Gene List
ICR-geneset Gene List. IMAGE ID UniGene Locus Name Cluster 20115 Hs.62185 SLC9A6 solute carrier family 9 (sodium/hydrogen exchanger), isoform 6 21738 21899 Hs.78353 SRPK2 SFRS protein kinase 2 21908 Hs.79133 CDH8 cadherin 8, type 2 22040 Hs.151738 MMP9 matrix metalloproteinase 9 (gelatinase B, 92kD gelatinase, 92kD type IV collagenase) 22411 Hs.183 FY Duffy blood group 22731 Hs.1787 PHRET1 PH domain containing protein in retina 1 22859 Hs.356487 ESTs 22883 Hs.150926 FPGT fucose-1-phosphate guanylyltransferase 22918 Hs.346868 EBNA1BP2 EBNA1 binding protein 2 23012 Hs.158205 BLZF1 basic leucine zipper nuclear factor 1 (JEM-1) 23073 Hs.284244 FGF2 fibroblast growth factor 2 (basic) 23173 Hs.151051 MAPK10 mitogen-activated protein kinase 10 23185 Hs.289114 TNC tenascin C (hexabrachion) 23282 Hs.8024 IK IK cytokine, down-regulator of HLA II 23353 23431 Hs.50421 RB1CC1 RB1-inducible coiled-coil 1 23514 23548 Hs.71848 Human clone 23548 mRNA sequence 23629 Hs.135587 Human clone 23629 mRNA sequence 23658 Hs.265855 SETMAR SET domain and mariner transposase fusion gene 23676 Hs.100841 Homo sapiens clone 23676 mRNA sequence 23772 Hs.78788 LZTR1 leucine-zipper-like transcriptional regulator, 1 23776 Hs.75438 QDPR quinoid dihydropteridine reductase 23804 Hs.343586 ZFP36 zinc finger protein 36, C3H type, homolog (mouse) 23831 Hs.155247 ALDOC aldolase C, fructose-bisphosphate 23878 Hs.99902 OPCML opioid binding protein/cell adhesion molecule-like 23903 Hs.12526 Homo sapiens clone 23903 mRNA sequence 23932 Hs.368063 Human clone 23932 mRNA sequence 24004 -

Contig Protein Description Symbol Anterior Posterior Ratio
Table S2. List of proteins detected in anterior and posterior intestine pooled samples. Data on protein expression are mean ± SEM of 4 pools fed the experimental diets. The number of the contig in the Sea Bream Database (http://nutrigroup-iats.org/seabreamdb) is indicated. Contig Protein Description Symbol Anterior Posterior Ratio Ant/Pos C2_6629 1,4-alpha-glucan-branching enzyme GBE1 0.88±0.1 0.91±0.03 0.98 C2_4764 116 kDa U5 small nuclear ribonucleoprotein component EFTUD2 0.74±0.09 0.71±0.05 1.03 C2_299 14-3-3 protein beta/alpha-1 YWHAB 1.45±0.23 2.18±0.09 0.67 C2_268 14-3-3 protein epsilon YWHAE 1.28±0.2 2.01±0.13 0.63 C2_2474 14-3-3 protein gamma-1 YWHAG 1.8±0.41 2.72±0.09 0.66 C2_1017 14-3-3 protein zeta YWHAZ 1.33±0.14 4.41±0.38 0.30 C2_34474 14-3-3-like protein 2 YWHAQ 1.3±0.11 1.85±0.13 0.70 C2_4902 17-beta-hydroxysteroid dehydrogenase 14 HSD17B14 0.93±0.05 2.33±0.09 0.40 C2_3100 1-acylglycerol-3-phosphate O-acyltransferase ABHD5 ABHD5 0.85±0.07 0.78±0.13 1.10 C2_15440 1-phosphatidylinositol phosphodiesterase PLCD1 0.65±0.12 0.4±0.06 1.65 C2_12986 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 PLCD1 0.76±0.08 1.15±0.16 0.66 C2_4412 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 PLCG2 1.13±0.08 2.08±0.27 0.54 C2_3170 2,4-dienoyl-CoA reductase, mitochondrial DECR1 1.16±0.1 0.83±0.03 1.39 C2_1520 26S protease regulatory subunit 10B PSMC6 1.37±0.21 1.43±0.04 0.96 C2_4264 26S protease regulatory subunit 4 PSMC1 1.2±0.2 1.78±0.08 0.68 C2_1666 26S protease regulatory subunit 6A PSMC3 1.44±0.24 1.61±0.08 -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

(CS-ⅣA-Be), a Novel IL-6R Antagonist, Inhibits IL-6/STAT3
Author Manuscript Published OnlineFirst on February 29, 2016; DOI: 10.1158/1535-7163.MCT-15-0551 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Chikusetsusaponin Ⅳa butyl ester (CS-Ⅳa-Be), a novel IL-6R antagonist, inhibits IL-6/STAT3 signaling pathway and induces cancer cell apoptosis Jie Yang 1, 2, Shihui Qian 2, Xueting Cai 1, 2, Wuguang Lu 1, 2, Chunping Hu 1, 2, * Xiaoyan Sun1, 2, Yang Yang1, 2, Qiang Yu 3, S. Paul Gao 4, Peng Cao 1, 2 1. Affiliated Hospital of Integrated Traditional Chinese and Western Medicine, Nanjing University of Chinese Medicine, Nanjing 210028, China 2. Laboratory of Cellular and Molecular Biology, Jiangsu Province Academy of Traditional Chinese Medicine, Nanjing 210028, China 3. Shanghai Institute of Materia Medical, Chinese Academy of Sciences, Shanghai, 201203, China 4. Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, NY10065, USA Running title: CS-Ⅳa-Be, a novel IL-6R antagonist, inhibits IL-6/STAT3 Keywords: Chikusetsusaponin Ⅳ a butyl ester (CS- Ⅳ a-Be), STAT3, IL-6R, antagonist, cancer Grant support: P. Cao received Jiangsu Province Funds for Distinguished Young Scientists (BK20140049) grant, J. Yang received National Natural Science Foundation of China (No. 81403151) grant, and X.Y. Sun received National Natural Science Foundation of China (No. 81202576) grant. Corresponding author: Peng Cao Institute: Laboratory of Cellular and Molecular Biology, Jiangsu Province Academy of Traditional Chinese Medicine, Nanjing 210028, Jiangsu, China Mailing address: 100#, Shizi Street, Hongshan Road, Nanjing, Jiangsu, China Tel: +86-25-85608666 Fax: +86-25-85608666 Email address: [email protected] The first co-authors: Jie Yang and Shihui Qian The authors disclose no potential conflicts of interest. -
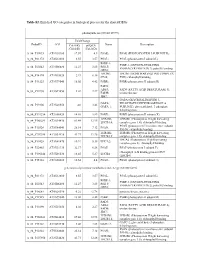
Table S2. Enriched GO Categories in Biological Process for the Shared Degs
Table S2. Enriched GO categories in biological process for the shared DEGs photosynthesis (GO ID:15979) Fold Change ProbeID AGI Col-0(R) pifQ(D) Name Description /Col-0(D) /Col-0(D) A_84_P19035 AT1G30380 17.07 4.9 PSAK; PSAK (PHOTOSYSTEM I SUBUNIT K) A_84_P21372 AT4G12800 8.55 3.57 PSAL; PSAL (photosystem I subunit L) PSBP-1; PSBP-1 (OXYGEN-EVOLVING A_84_P20343 AT1G06680 12.27 3.85 PSII-P; ENHANCER PROTEIN 2); poly(U) binding OEE2; LHCB6; LHCB6 (LIGHT HARVESTING COMPLEX A_84_P14174 AT1G15820 23.9 6.16 CP24; PSII); chlorophyll binding A_84_P11525 AT1G79040 16.02 4.42 PSBR; PSBR (photosystem II subunit R) FAD5; ADS3; FAD5 (FATTY ACID DESATURASE 5); A_84_P19290 AT3G15850 4.02 2.27 FADB; oxidoreductase JB67; GAPA (GLYCERALDEHYDE 3- GAPA; PHOSPHATE DEHYDROGENASE A A_84_P19306 AT3G26650 4.6 3.43 GAPA-1; SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase A_84_P193234 AT2G06520 14.01 3.89 PSBX; PSBX (photosystem II subunit X) LHB1B1; LHB1B1 (Photosystem II light harvesting A_84_P160283 AT2G34430 89.44 32.95 LHCB1.4; complex gene 1.4); chlorophyll binding PSAN (photosystem I reaction center subunit A_84_P10324 AT5G64040 26.14 7.12 PSAN; PSI-N); calmodulin binding LHB1B2; LHB1B2 (Photosystem II light harvesting A_84_P207958 AT2G34420 41.71 12.26 LHCB1.5; complex gene 1.5); chlorophyll binding LHCA2 (Photosystem I light harvesting A_84_P19428 AT3G61470 10.91 5.36 LHCA2; complex gene 2); chlorophyll binding A_84_P22465 AT1G31330 32.37 6.58 PSAF; PSAF (photosystem I subunit F) chlorophyll A-B binding protein CP29 A_84_P190244 AT5G01530 16.45 5.27 LHCB4 -

Progress in the Field of Aspartic Proteinases in Cheese Manufacturing
Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering Sirma Yegin, Peter Dekker To cite this version: Sirma Yegin, Peter Dekker. Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering. Dairy Science & Technology, EDP sciences/Springer, 2013, 93 (6), pp.565-594. 10.1007/s13594-013-0137-2. hal-01201447 HAL Id: hal-01201447 https://hal.archives-ouvertes.fr/hal-01201447 Submitted on 17 Sep 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Dairy Sci. & Technol. (2013) 93:565–594 DOI 10.1007/s13594-013-0137-2 REVIEW PAPER Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering Sirma Yegin & Peter Dekker Received: 25 February 2013 /Revised: 16 May 2013 /Accepted: 21 May 2013 / Published online: 27 June 2013 # INRA and Springer-Verlag France 2013 Abstract Aspartic proteinases are an important class of proteinases which are widely used as milk-coagulating agents in industrial cheese production. They are available from a wide range of sources including mammals, plants, and microorganisms. -

The Nutrition and Food Web Archive Medical Terminology Book
The Nutrition and Food Web Archive Medical Terminology Book www.nafwa. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Fatty Acid Biosynthesis
BI/CH 422/622 ANABOLISM OUTLINE: Photosynthesis Carbon Assimilation – Calvin Cycle Carbohydrate Biosynthesis in Animals Gluconeogenesis Glycogen Synthesis Pentose-Phosphate Pathway Regulation of Carbohydrate Metabolism Anaplerotic reactions Biosynthesis of Fatty Acids and Lipids Fatty Acids contrasts Diversification of fatty acids location & transport Eicosanoids Synthesis Prostaglandins and Thromboxane acetyl-CoA carboxylase Triacylglycerides fatty acid synthase ACP priming Membrane lipids 4 steps Glycerophospholipids Control of fatty acid metabolism Sphingolipids Isoprene lipids: Cholesterol ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1 ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1. Biosynthesis of fatty acids 2. Regulation of fatty acid degradation and synthesis 3. Assembly of fatty acids into triacylglycerol and phospholipids 4. Metabolism of isoprenes a. Ketone bodies and Isoprene biosynthesis b. Isoprene polymerization i. Cholesterol ii. Steroids & other molecules iii. Regulation iv. Role of cholesterol in human disease ANABOLISM II: Biosynthesis of Fatty Acids & Lipids Lipid Fat Biosynthesis Catabolism Fatty Acid Fatty Acid Degradation Synthesis Ketone body Isoprene Utilization Biosynthesis 2 Catabolism Fatty Acid Biosynthesis Anabolism • Contrast with Sugars – Lipids have have hydro-carbons not carbo-hydrates – more reduced=more energy – Long-term storage vs short-term storage – Lipids are essential for structure in ALL organisms: membrane phospholipids • Catabolism of fatty acids –produces acetyl-CoA –produces reducing -

Expression Analysis and Functional Significance of Chondroitin Sulphate Proteoglycans and Heparan Sulphate Proteoglycans in Prostate Cancer
EXPRESSION ANALYSIS AND FUNCTIONAL SIGNIFICANCE OF CHONDROITIN SULPHATE PROTEOGLYCANS AND HEPARAN SULPHATE PROTEOGLYCANS IN PROSTATE CANCER TENG HUI FANG, YVONNE (BSc, Hons) NATIONAL UNIVERSITY OF SINGAPORE A THESIS SUBMITTED FOR THE DEGREE OF DOCTOR OF PHILOSOPHY DEPARTMENT OF ANATOMY YONG LOO LIN SCHOOL OF MEDICINE NATIONAL UNIVERSITY OF SINGAPORE 2010 Acknowledgements ACKNOWLEDGEMENTS My PhD candidature would not have been easier, if not for the help, guidance and support from many mentors, family, friends and colleagues. My utmost gratitude goes to my mentor, Associate Professor George Yip Wai Cheong, for his unfailing guidance, patience and trust in me. Through him, I have learnt much, in terms of scientific skills and knowledge acquisition. In addition, I have benefitted from his many interesting stories that have also made my candidature a very enjoyable one indeed. The most important lesson that I have learnt from Dr George Yip would be the importance of thinking critically, a skill that is not only important in the scientific arena, but also one that applies to our everyday life. I would also like to thank my co-supervisor, Associate Professor Chia Sing Joo, for his support and advice. His comments have always added an interesting clinical perspective to the type of scientific research that I have been working on during my candidature. My deepest appreciation goes to Professor Bay Boon Huat for his timely advice and support. Professor Bay never fails to think for our welfare and has always shown genuine concern for us in all aspects of our life. He has taught us the importance of forming great relationships, one that supersedes any material wealth and status.