Identification and Functional Characterization of Adipogenesis-Related Genes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cloud Fonts in Microsoft Office
APRIL 2019 Guide to Cloud Fonts in Microsoft® Office 365® Cloud fonts are available to Office 365 subscribers on all platforms and devices. Documents that use cloud fonts will render correctly in Office 2019. Embed cloud fonts for use with older versions of Office. Reference article from Microsoft: Cloud fonts in Office DESIGN TO PRESENT Terberg Design, LLC Index MICROSOFT OFFICE CLOUD FONTS A B C D E Legend: Good choice for theme body fonts F G H I J Okay choice for theme body fonts Includes serif typefaces, K L M N O non-lining figures, and those missing italic and/or bold styles P R S T U Present with most older versions of Office, embedding not required V W Symbol fonts Language-specific fonts MICROSOFT OFFICE CLOUD FONTS Abadi NEW ABCDEFGHIJKLMNOPQRSTUVWXYZ abcdefghijklmnopqrstuvwxyz 01234567890 Abadi Extra Light ABCDEFGHIJKLMNOPQRSTUVWXYZ abcdefghijklmnopqrstuvwxyz 01234567890 Note: No italic or bold styles provided. Agency FB MICROSOFT OFFICE CLOUD FONTS ABCDEFGHIJKLMNOPQRSTUVWXYZ abcdefghijklmnopqrstuvwxyz 01234567890 Agency FB Bold ABCDEFGHIJKLMNOPQRSTUVWXYZ abcdefghijklmnopqrstuvwxyz 01234567890 Note: No italic style provided Algerian MICROSOFT OFFICE CLOUD FONTS ABCDEFGHIJKLMNOPQRSTUVWXYZ 01234567890 Note: Uppercase only. No other styles provided. Arial MICROSOFT OFFICE CLOUD FONTS ABCDEFGHIJKLMNOPQRSTUVWXYZ abcdefghijklmnopqrstuvwxyz 01234567890 Arial Italic ABCDEFGHIJKLMNOPQRSTUVWXYZ abcdefghijklmnopqrstuvwxyz 01234567890 Arial Bold ABCDEFGHIJKLMNOPQRSTUVWXYZ abcdefghijklmnopqrstuvwxyz 01234567890 Arial Bold Italic ABCDEFGHIJKLMNOPQRSTUVWXYZ -

Combined Human Epididymis 4 and Carbohydrate Antigen 125 Serum Protein Levels Diagnostic Value in Ovarian Cancer
ISSN: 2640-5180 Madridge Journal of Cancer Study & Research Research Article Open Access Combined Human Epididymis 4 and Carbohydrate Antigen 125 Serum Protein Levels Diagnostic value in Ovarian Cancer Salwa Hassan Teama1*, Reham El Shimy2 and Hebatallah Gamal3 1Department of Molecular Biology, Medical Ain Shams Research Institute (MASRI), Faculty of Medicine, Ain Shams University, Cairo, Egypt 2Department of Clinical and Chemical Pathology, National Cancer Institute, Faculty of Medicine, Cairo University, Cairo, Egypt 3Departement of Surgical Oncology, National Cancer Institute, Faculty of Medicine, Cairo University, Cairo, Egypt Article Info Abstract *Corresponding author: Objective: Human Epididymis 4 (HE-4) protein, a new candidate for ovarian cancer Salwa Hassan Teama detection shows promising diagnostic value for ovarian cancer diagnosis, this study aimed Department of Molecular Biology Medical Ain Shams Research Institute to assess the diagnostic significance of combined Human Epididymis 4 and Carbohydrate (MASRI) Antigen 125 (CA-125) serum protein levels in ovarian cancer detection. Faculty of Medicine Ain Shams University Subjects and Methods: A clinical case control study include; forty nine subjects; patients Abbasia, Cairo with ovarian cancer (n=33), non-cancer control group (n=16). Serum protein levels of Egypt HE-4 were measured using an enzyme linked immune sorbent assay (ELISA). All data were Tel: 0020-1005293116 E-mail: [email protected] analyzed by SPSS software (version 21.0.0; IPM SPSS, Chicago, IL, USA). Results: The results showed that increased serum protein concentration of HE-4 (pMol/L) and Received: July 25, 2018 CA-125 (U/ml) in the ovarian cancer group mean (SD)/median (range) 329.61±336.55/199 Accepted: August 13, 2018 Published: August 17, 2018 (28.72-1064) and 521.36±572.60/287 (10.50-2377), than non-cancer control group 64.80±38.51/54.53 (21 -160) and 28.35±10.80/28 (10-50) respectively (p<0.05). -

WFDC2 Control Peptide
AP10066CP-N OriGene Technologies Inc. OriGene EU Acris Antibodies GmbH 9620 Medical Center Drive, Ste 200 Schillerstr. 5 Rockville, MD 20850 32052 Herford UNITED STATES GERMANY Phone: +1-888-267-4436 Phone: +49-5221-34606-0 Fax: +1-301-340-8606 Fax: +49-5221-34606-11 [email protected] [email protected] WFDC2 control peptide Alternate names: ESPE4, Epidydimal Secretory Protein E4, HE4, Major Epididymis-Specific protein E4, WAP four-disulfide core domain protein 2, WAP5, WFDC2 Catalog No.: AP10066CP-N Quantity: 0.25 mg Concentration: 2.5 mg/ml Background: The whey acidic protein (WAP) domain is a conserved motif, containing eight cysteines found in a characteristic 4-disulphide core arrangement, that is present in a number of otherwise unrelated proteins. One of these proteins, which contains two WAP domains, is HE4 (also known as WFDC2), originally described as an epididymis specific protein but more recently suggested to be a putative serum tumour marker for ovarian cancer and a presumptive role in natural immunity. The HE4 protein expression is not only confined to epidydimis but is expressed in a number of normal human tissues out side the reproductive system, including regions of the respiratory tract and nasopharynx and in a subset of lung tumor cell lines. HE4 gene expression was highest in normal human trachea and salivary gland, and to a lesser extent, lung, prostate, pituitary gland, thyroid, and kidney. Highest level of expression of ESPE4 was observed in adenocarcinomas of the lung, and occasional breast, transitional cell and pancreatic carcinomas (1). The WFDC2 gene under goes extensive splicing in malignant tissues that give rise to five WAP domain containing iso forms (2). -

Contig Protein Description Symbol Anterior Posterior Ratio
Table S2. List of proteins detected in anterior and posterior intestine pooled samples. Data on protein expression are mean ± SEM of 4 pools fed the experimental diets. The number of the contig in the Sea Bream Database (http://nutrigroup-iats.org/seabreamdb) is indicated. Contig Protein Description Symbol Anterior Posterior Ratio Ant/Pos C2_6629 1,4-alpha-glucan-branching enzyme GBE1 0.88±0.1 0.91±0.03 0.98 C2_4764 116 kDa U5 small nuclear ribonucleoprotein component EFTUD2 0.74±0.09 0.71±0.05 1.03 C2_299 14-3-3 protein beta/alpha-1 YWHAB 1.45±0.23 2.18±0.09 0.67 C2_268 14-3-3 protein epsilon YWHAE 1.28±0.2 2.01±0.13 0.63 C2_2474 14-3-3 protein gamma-1 YWHAG 1.8±0.41 2.72±0.09 0.66 C2_1017 14-3-3 protein zeta YWHAZ 1.33±0.14 4.41±0.38 0.30 C2_34474 14-3-3-like protein 2 YWHAQ 1.3±0.11 1.85±0.13 0.70 C2_4902 17-beta-hydroxysteroid dehydrogenase 14 HSD17B14 0.93±0.05 2.33±0.09 0.40 C2_3100 1-acylglycerol-3-phosphate O-acyltransferase ABHD5 ABHD5 0.85±0.07 0.78±0.13 1.10 C2_15440 1-phosphatidylinositol phosphodiesterase PLCD1 0.65±0.12 0.4±0.06 1.65 C2_12986 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 PLCD1 0.76±0.08 1.15±0.16 0.66 C2_4412 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 PLCG2 1.13±0.08 2.08±0.27 0.54 C2_3170 2,4-dienoyl-CoA reductase, mitochondrial DECR1 1.16±0.1 0.83±0.03 1.39 C2_1520 26S protease regulatory subunit 10B PSMC6 1.37±0.21 1.43±0.04 0.96 C2_4264 26S protease regulatory subunit 4 PSMC1 1.2±0.2 1.78±0.08 0.68 C2_1666 26S protease regulatory subunit 6A PSMC3 1.44±0.24 1.61±0.08 -

CLASP2 Antibody Product Type
PRODUCT INFORMATION Product name: CLASP2 antibody Product type: Primary antibodies Description: Rabbit polyclonal to CLASP2 Immunogen:3 synthetic peptides (human) conjugated to KLH Reacts with:Hu, Ms Tested applications:ELISA, WB and IF GENE INFORMATION Gene Symbol: CLASP2 Gene Name:cytoplasmic linker associated protein 2 Ensembl ID:ENSG00000163539 Entrez GeneID:23122 GenBank Accession number:AB014527 Swiss-Prot:O75122 Molecular weight of CLASP2: 165.9 & 108.6kDa Function:Microtubule plus-end tracking protein that promotes the stabilization of dynamic microtubules. Involved in the nucleation of noncentrosomal microtubules originating from the trans-Golgi network (TGN). Required for the polarization of the cytoplasmic microtubule arrays in migrating cells towards the leading edge of the cell. May act at the cell cortex to enhance the frequency of rescue of depolymerizing microtubules by attaching their plus- ends to cortical platforms composed of ERC1 and PHLDB2. This cortical microtubule stabilizing activity is regulated at least in part by phosphatidylinositol 3-kinase signaling. Also performs a similar stabilizing function at the kinetochore which is essential for the bipolar alignment of chromosomes on the mitotic spindle. Acts as a mediator of ERBB2- dependent stabilization of microtubules at the cell cortex. Expected subcellular localization:Cytoplasm › cytoskeleton. Cytoplasm › cytoskeleton › microtubule organizing center › centrosome. Chromosome › centromere › kinetochore. Cytoplasm › cytoskeleton › spindle. Golgi apparatus. Golgi apparatus › trans-Golgi network. Cell membrane. Cell projection › ruffle membrane. Note: Localizes to microtubule plus ends. Localizes to centrosomes, kinetochores and the mitotic spindle from prometaphase. Subsequently localizes to the spindle midzone from anaphase and to the midbody from telophase. In migrating cells localizes to the plus ends of microtubules within the cell body and to the entire microtubule lattice within the lamella. -
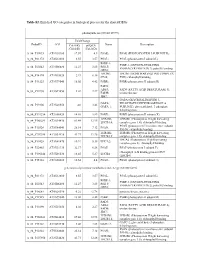
Table S2. Enriched GO Categories in Biological Process for the Shared Degs
Table S2. Enriched GO categories in biological process for the shared DEGs photosynthesis (GO ID:15979) Fold Change ProbeID AGI Col-0(R) pifQ(D) Name Description /Col-0(D) /Col-0(D) A_84_P19035 AT1G30380 17.07 4.9 PSAK; PSAK (PHOTOSYSTEM I SUBUNIT K) A_84_P21372 AT4G12800 8.55 3.57 PSAL; PSAL (photosystem I subunit L) PSBP-1; PSBP-1 (OXYGEN-EVOLVING A_84_P20343 AT1G06680 12.27 3.85 PSII-P; ENHANCER PROTEIN 2); poly(U) binding OEE2; LHCB6; LHCB6 (LIGHT HARVESTING COMPLEX A_84_P14174 AT1G15820 23.9 6.16 CP24; PSII); chlorophyll binding A_84_P11525 AT1G79040 16.02 4.42 PSBR; PSBR (photosystem II subunit R) FAD5; ADS3; FAD5 (FATTY ACID DESATURASE 5); A_84_P19290 AT3G15850 4.02 2.27 FADB; oxidoreductase JB67; GAPA (GLYCERALDEHYDE 3- GAPA; PHOSPHATE DEHYDROGENASE A A_84_P19306 AT3G26650 4.6 3.43 GAPA-1; SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase A_84_P193234 AT2G06520 14.01 3.89 PSBX; PSBX (photosystem II subunit X) LHB1B1; LHB1B1 (Photosystem II light harvesting A_84_P160283 AT2G34430 89.44 32.95 LHCB1.4; complex gene 1.4); chlorophyll binding PSAN (photosystem I reaction center subunit A_84_P10324 AT5G64040 26.14 7.12 PSAN; PSI-N); calmodulin binding LHB1B2; LHB1B2 (Photosystem II light harvesting A_84_P207958 AT2G34420 41.71 12.26 LHCB1.5; complex gene 1.5); chlorophyll binding LHCA2 (Photosystem I light harvesting A_84_P19428 AT3G61470 10.91 5.36 LHCA2; complex gene 2); chlorophyll binding A_84_P22465 AT1G31330 32.37 6.58 PSAF; PSAF (photosystem I subunit F) chlorophyll A-B binding protein CP29 A_84_P190244 AT5G01530 16.45 5.27 LHCB4 -

Mouse Phldb2 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Phldb2 Knockout Project (CRISPR/Cas9) Objective: To create a Phldb2 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Phldb2 gene (NCBI Reference Sequence: NM_001252442 ; Ensembl: ENSMUSG00000033149 ) is located on Mouse chromosome 16. 19 exons are identified, with the ATG start codon in exon 2 and the TAG stop codon in exon 19 (Transcript: ENSMUST00000076333). Exon 2 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for a conditional allele activated in neurons exhibit impaired LTP. Exon 2 starts from the coding region. Exon 2 covers 33.87% of the coding region. The size of effective KO region: ~1337 bp. The KO region does not have any other known gene. Page 1 of 9 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 19 Legends Exon of mouse Phldb2 Knockout region Page 2 of 9 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 2 is aligned with itself to determine if there are tandem repeats. Tandem repeats are found in the dot plot matrix. The gRNA site is selected outside of these tandem repeats. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section downstream of Exon 2 is aligned with itself to determine if there are tandem repeats. -

Fatty Acid Biosynthesis
BI/CH 422/622 ANABOLISM OUTLINE: Photosynthesis Carbon Assimilation – Calvin Cycle Carbohydrate Biosynthesis in Animals Gluconeogenesis Glycogen Synthesis Pentose-Phosphate Pathway Regulation of Carbohydrate Metabolism Anaplerotic reactions Biosynthesis of Fatty Acids and Lipids Fatty Acids contrasts Diversification of fatty acids location & transport Eicosanoids Synthesis Prostaglandins and Thromboxane acetyl-CoA carboxylase Triacylglycerides fatty acid synthase ACP priming Membrane lipids 4 steps Glycerophospholipids Control of fatty acid metabolism Sphingolipids Isoprene lipids: Cholesterol ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1 ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1. Biosynthesis of fatty acids 2. Regulation of fatty acid degradation and synthesis 3. Assembly of fatty acids into triacylglycerol and phospholipids 4. Metabolism of isoprenes a. Ketone bodies and Isoprene biosynthesis b. Isoprene polymerization i. Cholesterol ii. Steroids & other molecules iii. Regulation iv. Role of cholesterol in human disease ANABOLISM II: Biosynthesis of Fatty Acids & Lipids Lipid Fat Biosynthesis Catabolism Fatty Acid Fatty Acid Degradation Synthesis Ketone body Isoprene Utilization Biosynthesis 2 Catabolism Fatty Acid Biosynthesis Anabolism • Contrast with Sugars – Lipids have have hydro-carbons not carbo-hydrates – more reduced=more energy – Long-term storage vs short-term storage – Lipids are essential for structure in ALL organisms: membrane phospholipids • Catabolism of fatty acids –produces acetyl-CoA –produces reducing -

Saturated Long-Chain Fatty Acid-Producing Bacteria Contribute
Zhao et al. Microbiome (2018) 6:107 https://doi.org/10.1186/s40168-018-0492-6 RESEARCH Open Access Saturated long-chain fatty acid-producing bacteria contribute to enhanced colonic motility in rats Ling Zhao1†, Yufen Huang2†, Lin Lu1†, Wei Yang1, Tao Huang1, Zesi Lin3, Chengyuan Lin1,4, Hiuyee Kwan1, Hoi Leong Xavier Wong1, Yang Chen5, Silong Sun2, Xuefeng Xie2, Xiaodong Fang2,5, Huanming Yang6, Jian Wang6, Lixin Zhu7* and Zhaoxiang Bian1* Abstract Background: The gut microbiota is closely associated with gastrointestinal (GI) motility disorder, but the mechanism(s) by which bacteria interact with and affect host GI motility remains unclear. In this study, through using metabolomic and metagenomic analyses, an animal model of neonatal maternal separation (NMS) characterized by accelerated colonic motility and gut dysbiosis was used to investigate the mechanism underlying microbiota-driven motility dysfunction. Results: An excess of intracolonic saturated long-chain fatty acids (SLCFAs) was associated with enhanced bowel motility in NMS rats. Heptadecanoic acid (C17:0) and stearic acid (C18:0), as the most abundant odd- and even- numbered carbon SLCFAs in the colon lumen, can promote rat colonic muscle contraction and increase stool frequency. Increase of SLCFAs was positively correlated with elevated abundances of Prevotella, Lactobacillus, and Alistipes. Functional annotation found that the level of bacterial LCFA biosynthesis was highly enriched in NMS group. Essential synthetic genes Fabs were largely identified from the genera Prevotella, Lactobacillus, and Alistipes. Pseudo germ-free (GF) rats receiving fecal microbiota from NMS donors exhibited increased defecation frequency and upregulated bacterial production of intracolonic SLCFAs. Modulation of gut dysbiosis by neomycin effectively attenuated GI motility and reduced bacterial SLCFA generation in the colon lumen of NMS rats. -

Convergent Functional Genomics of Schizophrenia: from Comprehensive Understanding to Genetic Risk Prediction
Molecular Psychiatry (2012) 17, 887 -- 905 & 2012 Macmillan Publishers Limited All rights reserved 1359-4184/12 www.nature.com/mp IMMEDIATE COMMUNICATION Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction M Ayalew1,2,9, H Le-Niculescu1,9, DF Levey1, N Jain1, B Changala1, SD Patel1, E Winiger1, A Breier1, A Shekhar1, R Amdur3, D Koller4, JI Nurnberger1, A Corvin5, M Geyer6, MT Tsuang6, D Salomon7, NJ Schork7, AH Fanous3, MC O’Donovan8 and AB Niculescu1,2 We have used a translational convergent functional genomics (CFG) approach to identify and prioritize genes involved in schizophrenia, by gene-level integration of genome-wide association study data with other genetic and gene expression studies in humans and animal models. Using this polyevidence scoring and pathway analyses, we identify top genes (DISC1, TCF4, MBP, MOBP, NCAM1, NRCAM, NDUFV2, RAB18, as well as ADCYAP1, BDNF, CNR1, COMT, DRD2, DTNBP1, GAD1, GRIA1, GRIN2B, HTR2A, NRG1, RELN, SNAP-25, TNIK), brain development, myelination, cell adhesion, glutamate receptor signaling, G-protein-- coupled receptor signaling and cAMP-mediated signaling as key to pathophysiology and as targets for therapeutic intervention. Overall, the data are consistent with a model of disrupted connectivity in schizophrenia, resulting from the effects of neurodevelopmental environmental stress on a background of genetic vulnerability. In addition, we show how the top candidate genes identified by CFG can be used to generate a genetic risk prediction score (GRPS) to aid schizophrenia diagnostics, with predictive ability in independent cohorts. The GRPS also differentiates classic age of onset schizophrenia from early onset and late-onset disease. -

Long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression
RESEARCH ARTICLE Long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression Yichi Xu1,2, Weimin Guo1, Ping Li3, Yan Zhang3, Meng Zhao1,4, Zenghua Fan1,2, Zhihu Zhao3, Jun Yan1,4* 1 CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China, 2 University of Chinese Academy of Sciences, Shanghai, China, 3 Beijing Institute of Biotechnology, Beijing, China, 4 Institute of Neuroscience, State Key Laboratory a11111 of Neuroscience, CAS Center for Excellence in Brain Science, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China * [email protected] Abstract OPEN ACCESS Mammalian circadian rhythm is established by the negative feedback loops consisting of a Citation: Xu Y, Guo W, Li P, Zhang Y, Zhao M, Fan Z, et al. (2016) Long-Range Chromosome Interactions set of clock genes, which lead to the circadian expression of thousands of downstream Mediated by Cohesin Shape Circadian Gene genes in vivo. As genome-wide transcription is organized under the high-order chromosome Expression. PLoS Genet 12(5): e1005992. structure, it is largely uncharted how circadian gene expression is influenced by chromo- doi:10.1371/journal.pgen.1005992 some architecture. We focus on the function of chromatin structure proteins cohesin as well — Editor: Achim Kramer, Charité Universitätsmedizin as CTCF (CCCTC-binding factor) in circadian rhythm. Using circular chromosome confor- Berlin, GERMANY mation capture sequencing, we systematically examined the interacting loci of a Bmal1- Received: October 10, 2015 bound super-enhancer upstream of a clock gene Nr1d1 in mouse liver. These interactions Accepted: March 27, 2016 are largely stable in the circadian cycle and cohesin binding sites are enriched in the interac- Published: May 2, 2016 tome. -

Ultimate++ Forum It Higher Priority Now
Subject: It's suspected to be an issue with Font. Posted by Lance on Fri, 18 Mar 2011 22:50:28 GMT View Forum Message <> Reply to Message The programs I used to compare are UWord from the UPP, and MS Word. The platform is Windows 7. The text I used to test is: The problem with U++ drawed text is that some characters are notably larger than others and some have incorrect horizontal displacement. Please see attached picture for a visual effect. I also encountered issue where chinese characters are displayed correctly displayed on Windows but are blank on Ubuntu. And when I copies the same text that was displayed as blank to, say gedit, the text displayed correctly as in Windows. That part I will attach picture in future. File Attachments 1) font problem.png, downloaded 650 times Subject: Re: It's suspected to be an issue with Font. Posted by mirek on Sun, 10 Apr 2011 12:42:52 GMT View Forum Message <> Reply to Message Lance wrote on Fri, 18 March 2011 18:50The programs I used to compare are UWord from the UPP, and MS Word. The platform is Windows 7. The text I used to test is: The problem with U++ drawed text is that some characters are notably larger than others and some have incorrect horizontal displacement. It works fine on my Windows 7. However, I believe that the problem is caused by font substitution mechanism and perhaps on your system, you have some font that takes precendence for some glyphs, but does not contain other characters.