Assessing the 5S Ribosomal RNA Heterogeneity in Arabidopsis Thaliana Using Short RNA Next Generation Sequencing Data Maciej Szymanski* and Wojciech M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Organization and Evolution of 5S Ribosomal Dna in the Fish Genome
In: Focus on Genome Research ISSN 1-59033-960-6 Editor: Clyde R. Williams, pp335-363 ©2004 Nova Science Publishers, Inc. Chapter X ORGANIZATION AND EVOLUTION OF 5S RIBOSOMAL DNA IN THE FISH GENOME Cesar Martins 1 and Adriane Pinto Wasko 2 Departamento de Morfologia, Instituto de Biociências, Universidade Estadual Paulista, CEP 18618-000, Botucatu, SP, Brazil. Phone/Fax +55 14 38116264, 1e-mail [email protected]; 2e-mail: [email protected] ABSTRACT In higher eukaryotes, the 5S ribosomal multigene family (5S rDNA) is tandemly organized in repeat units composed of a coding region (5S rRNA gene) and a non-transcribed spacer sequence (NTS). Although the 5S rDNA organization has been investigated in several vertebrate species, present data are concentrated in mammals and amphibians, whereas other groups, such as fishes, have been poorly studied. To further the understanding on the dynamics and evolution of 5S rDNA arrays in the vertebrate genome, recent studies have focused on the genome organization of these sequences in fish species, which represent the base group of vertebrate evolution. It was evidenced that the chromosome distribution of the 5S rDNA is quite conserved among related fish species occupying an interstitial position in the chromosomes. Although the 5S rDNA clusters have been maintained conserved in the chromosomes, changes in the nucleotide sequences and organization of the repeat units have occurred in fish species, as demonstrated by the presence of 5S rDNA variant types within and between genomes clustered in distinct chromosome environments. These variants are distributed in two major classes, suggesting that such pattern could represent a primitive condition for the fish genome, as well as for vertebrates. -
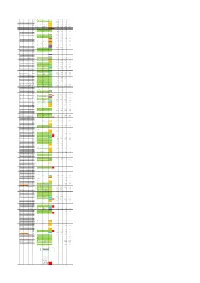
End Strand of Gene Gene Name Gene Function Starnd of Trascript
Genomic TSS strand Gene starnd of Transcipt relatively to TSS Start End Gene name coordinates relatively TSS type Control_fwd TEX_fwd Control_rev TEX_rev of gene function trascript gene group of TSS to ATG ribosomal 93 854 + rps12 + 1 protein S12 1 1 14.0638317 20.7539794 0 0 93 324 exon 1 + 111 rps12 intron 1 1 13.90756687 18.07149224 0.781323982 0.423550599 829 854 exon 2 + 496 rps12 exon2 2 24.22104343 15.24782157 0 0 30S 904 1371 + gene rps7 ribosomal + 2 protein S7 1303 -209 rps7 inter (ndhB) 2 20.47068832 9.035746118 0.625059185 0.847101199 NADH dehydrogen 1512 3535 + gene ndhB (ndh2) + ase subunit 3 2 1696 ndhB exon 1-inter 2 3.594090315 2.964854195 0.468794389 0.282367066 + 2209 ndhB exon 1-inter 2 46.09811492 4.09432246 0.468794389 0.423550599 1512 2237 exon 1 + 2756 ndhB exon 2 -inter 2 43.28534858 4.800240125 0.312529593 0.282367066 2756 3535 exon 2 + 3090 ndhB exon 2 -inter 2 17.50165719 15.95373924 0.312529593 0 + 3192 ndhB exon 2 -inter 2 140.6383167 117.6058831 2.812766334 1.694202397 - 3462 ndhB exon 2 -inter 3 1.406383167 1.129468265 1.406383167 3.67077186 4 3633 3712 + tRNA tRNA-Leu-CAA + 3610 -23 tRNA-Leu 1 77.19480938 84.85130339 0.625059185 0 + 3633 1 tRNA-Leu 1 359.5652963 649.0207016 0.781323982 0 photosyste 3954 4058 + gene psbM m II protein + 5 M 3775 -179 psbM 2 20.47068832 12.00060031 0 0.141183533 + 3954 psbM-0 2 69.22530477 28.37789015 0.156264796 0 hypothetical 4182 5073 + gene ycf66 6 protein 4182 4287 exon 1 4772 5073 exon 2 7 5202 5113 - gene ycf (ORF29) - 5299 orf29 inter 1 0 0 3.125295926 3.67077186 -

HUMAN RIBOSOME BIOGENESIS and the REGULATION of the TUMOUR SUPPRESSOR P53
HUMAN RIBOSOME BIOGENESIS AND THE REGULATION OF THE TUMOUR SUPPRESSOR p53 Andria Pelava Submitted for Doctor of Philosophy Final submission: December 2016 Institute of Cell and Molecular Biosciences Faculty of Medical Sciences Newcastle University ii Abstract Ribosome production is an energetically expensive and, therefore, highly regulated process. Defects in ribosome biogenesis lead to genetic diseases called Ribosomopathies, such as Dyskeratosis Congenita (DC), and mutations in ribosomal proteins and ribosome biogenesis factors are linked to multiple types of cancer. During ribosome biogenesis, the ribosomal RNAs (rRNAs) are processed and modified, and defects in ribosome biogenesis lead to the activation of p53. This project aimed to investigate the functions of Dyskerin, mutated in X-linked DC, in human ribosome biogenesis and p53 regulation, and to explore the link between ribosome production and p53 homeostasis. Dyskerin is an rRNA pseudouridine synthase and required for telomere maintenance. There is some debate as to whether DC is the result of telomere maintenance or ribosome biogenesis defects. It is shown here that human Dyskerin is required for the production of both LSU and SSU, and knockdown of Dyskerin leads to p53 activation via inhibition of MDM2 via the 5S RNP, an LSU assembly intermediate which accumulates after ribosome biogenesis defects. My data indicate that p53 activation, due to defects in ribosome biogenesis, may contribute to the clinical symptoms seen in patients suffering with DC. In addition, it is shown that defects in early or late stages of SSU or LSU biogenesis, result in activation of p53 via the 5S RNP-MDM2 pathway, and that p53 is induced in less than 12 hours after ribosome biogenesis defects. -

Selenoprotein H Is an Essential Regulator of Redox Homeostasis That
Selenoprotein H is an essential regulator of redox PNAS PLUS homeostasis that cooperates with p53 in development and tumorigenesis Andrew G. Coxa, Allison Tsomidesa, Andrew J. Kima, Diane Saundersa, Katie L. Hwanga, Kimberley J. Evasonb, Jerry Heidelc, Kristin K. Brownd, Min Yuand, Evan C. Liend, Byung Cheon Leea,e, Sahar Nissima, Bryan Dickinsonf, Sagar Chhangawalag, Christopher J. Changh,i, John M. Asarad, Yariv Houvrasg, Vadim N. Gladysheva,j, and Wolfram Goesslinga,j,k,l,1 aBrigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115; bUniversity of Utah, Salt Lake City, UT 84112; cOregon State University, Corvallis, OR 97331; dBeth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02115; eKorea University, 02841 Seoul, Republic of Korea; fUniversity of Chicago, Chicago, IL 60637; gWeill Cornell Medical College and New York Presbyterian Hospital, New York, NY 10065; hHoward Hughes Medical Institute, Bethesda, MD 20815; iUniversity of California, Berkeley, CA 20815; jBroad Institute of MIT and Harvard, Cambridge, MA 02142; kHarvard Stem Cell Institute, Cambridge, MA 02138; and lDana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115 Edited by Leonard I. Zon, Howard Hughes Medical Institute, Boston Children’s Hospital, Harvard Medical School, Boston, MA, and accepted by Editorial Board Member Carol Prives July 18, 2016 (received for review January 7, 2016) Selenium, an essential micronutrient known for its cancer preven- mice exhibit impaired selenium transport from the liver to pe- tion properties, is incorporated into a class of selenocysteine- ripheral tissues and show growth retardation and impaired motor containing proteins (selenoproteins). Selenoprotein H (SepH) is a coordination (13, 14). -

UC Riverside Electronic Theses and Dissertations
UC Riverside UC Riverside Electronic Theses and Dissertations Title Characterization of AtRAP Function in Plant Immunity and in RNA Transportation Permalink https://escholarship.org/uc/item/8pk9k169 Author Wang, Huan Publication Date 2017 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA RIVERSIDE Characterization of AtRAP Function in Plant Immunity and in RNA Transportation A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Plant Pathology by Huan Wang December 2017 Dissertation Committee: Dr. Hailing Jin, Chairperson Dr. Julia Bailey-Serres Dr. Isgouhi Kaloshian Copyright by Huan Wang 2017 The Dissertation of Huan Wang is approved: Committee Chairperson University of California, Riverside ACKNOWLEDGEMENTS I would like to express my sincere gratitude and appreciation to all those who helped and encouraged me during my graduate study. My deepest gratitude goes first and foremost to Dr. Hailing Jin, my supervisor, for her patient and constant guidance and encouragement. No matter what the difficulty is, she always stands with me to give me very timely, experienced and smart advice to help me solve it quickly and efficiently. Her creative ideas, intelligent analysis, hardworking attitude and generous personality inspire and teach me a lot not only on my research but also on my philosophy of life. Secondly, I would like to express my heartfelt gratitude to my dissertation committee members, Dr. Julia Bailey-Serres and Dr. Isgouhi Kaloshian. I really appreciate your valuable suggestions and kindly help to my study. Dr. Julia Bailey-Serres continuously helps my study to be my guidance committee member, qualifying exam committee member, and dissertation committee member. -

Untersuchung Zur Rolle Des La-Verwandten Proteins LARP4B Im Mrna- Metabolismus
Untersuchung zur Rolle des La-verwandten Proteins LARP4B im mRNA-Metabolismus Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Julius-Maximilians-Universität Würzburg vorgelegt von Maritta Küspert aus Schwäbisch Hall Würzburg 2014 Eingereicht bei der Fakultät für Chemie und Pharmazie am: ....................................... Gutachter der schriftlichen Arbeit: 1. Gutachter: Prof. Dr. Utz Fischer 2. Gutachter: Prof. Dr. Alexander Buchberger Prüfer des öffentlichen Promotionskolloquiums: 1. Prüfer: Prof. Dr. Utz Fischer 2. Prüfer: Prof. Dr. Alexander Buchberger 3. Prüfer: Prof. Dr. Stefan Gaubatz Datum des öffentlichen Promotionskolloquiums: ......................................................... Doktorurkunde ausgehändigt am: ................................................................................ Zusammenfassung Eukaryotische messenger-RNAs (mRNAs) müssen diverse Prozessierungsreaktionen durchlaufen, bevor sie der Translationsmaschinerie als Template für die Proteinbiosynthese dienen können. Diese Reaktionen beginnen bereits kotranskriptionell und schließen das Capping, das Spleißen und die Polyadenylierung ein. Erst nach dem die Prozessierung abschlossen ist, kann die reife mRNA ins Zytoplasma transportiert und translatiert werden. mRNAs interagieren in jeder Phase ihres Metabolismus mit verschiedenen trans-agierenden Faktoren und bilden mRNA-Ribonukleoproteinkomplexe (mRNPs) aus. Dieser „mRNP-Code“ bestimmt das Schicksal jeder mRNA und reguliert dadurch die Genexpression auf posttranskriptioneller -

Supplemental Materials
The infection-tolerant mammalian reservoir of Lyme disease and other zoonoses broadly counters the inflammatory effects of endotoxin Supplemental Materials Figures S1-S5 Tables S1-S20 Figure S1. Digital photograph of two adult Peromyscus leucopus with exudative conjunctivitis and huddled together. The animals had received 10 mg/gm body of Escherichia coli lipopolysaccharide intraperitoneally the day before. Figure S2. Species- and tissue-specific responses to LPS. Independent differential gene expression analysis of RNA-seq data were performed for blood, spleen, and liver tissues of P. leucopus and M. musculus collected 4 h after injection with LPS or buffer alsone as control. These are represented as volcano plots with range- adjusted scales for the log2-transformed fold-changes on x-axes and log10-transformed FDR p values on y- axes. Colors of symbols denote the following: red, up-regulated gene with absolute fold-change > 4.0 and p value < 0.05; purple, down-regulated gene with absolute fold-change > 4.0 and p value < 0.05; and gray, all others. Numbers at the top left and right corners in each plot respresent numbers of down- and up-regulated genes, respectively. Figure 3 is same data with constant scales for x- and y-axes across the plots. Numerical values for each gene in the 6 datasets are provided in Tables S4-S9. Figure S3. Correlation of IL-10 and IL-10 P. leucopus and M. musculus from RNA-seq of spleen and of Figure 6B and TaBle S14. The scatter plot is log10 values of normalized unique reads of one coding sequence against another for each of the four groups, as defined in the legend for Figure 6 and indicated By different symBols. -

Specific Features of 5S Rrna Structure – Its Interactions with Macromolecules and Possible Functions
ISSN 0006-2979, Biochemistry (Moscow), 2008, Vol. 73, No. 13, pp. 1418-1437. © Pleiades Publishing, Ltd., 2008. Original Russian Text © A. V. Smirnov, N. S. Entelis, I. A. Krasheninnikov, R. Martin, I. A. Tarassov, 2008, published in Uspekhi Biologicheskoi Khimii, 2008, Vol. 48, pp. 133-180. REVIEW Specific Features of 5S rRNA Structure – Its Interactions with Macromolecules and Possible Functions A. V. Smirnov1,2*, N. S. Entelis2, I. A. Krasheninnikov1, R. Martin2, and I. A. Tarassov1 1Faculty of Biology, Lomonosov Moscow State University, 119991 Moscow, Russia; E-mail: [email protected] 2Department of Molecular and Cellular Genetics, UMR 7156, Centre National de la Recherche Scientifique – Universite Louis Pasteur, Strasbourg 67084, France Received November 19, 2007 Revision received February 18, 2008 Abstract—Small non-coding RNAs are today a topic of great interest for molecular biologists because they can be regarded as relicts of a hypothetical “RNA world” which, apparently, preceded the modern stage of organic evolution on Earth. The small molecule of 5S rRNA (~120 nucleotides) is a component of large ribosomal subunits of all living beings (5S rRNAs are not found only in mitoribosomes of fungi and metazoans). This molecule interacts with various protein factors and 23S (28S) rRNA. This review contains the accumulated data to date concerning 5S rRNA structure, interactions with other bio- logical macromolecules, intracellular traffic, and functions in the cell. DOI: 10.1134/S000629790813004X Key words: 5S rRNA, structure, RNA–protein interaction, ribosome, intracellular transport Small non-coding RNAs are now a topic of interest GENERAL INFORMATION ABOUT 5S rRNA for researchers as relicts of a hypothetical “RNA world” that apparently preceded the modern stage of organic 5S rRNA is a relatively small RNA molecule (~120 evolution on Earth. -

University of Groningen Genome Sequencing and Analysis of The
University of Groningen Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88 Pel, Herman J.; de Winde, Johannes H.; Archer, David B.; Dyer, Paul S.; Hofmann, Gerald; Schaap, Peter J.; Turner, Geoffrey; Albang, Richard; Albermann, Kaj; Andersen, Mikael R. Published in: Nature Biotechnology DOI: 10.1038/nbt1282 IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2007 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Pel, H. J., de Winde, J. H., Archer, D. B., Dyer, P. S., Hofmann, G., Schaap, P. J., Turner, G., Albang, R., Albermann, K., Andersen, M. R., Bendtsen, J. D., Benen, J. A. E., van den Berg, M., Breestraat, S., Caddick, M. X., Contreras, R., Cornell, M., Coutinho, P. M., Danchin, E. G. J., ... de Vries, R. P. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nature Biotechnology, 25(2), 221-231. https://doi.org/10.1038/nbt1282 Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). The publication may also be distributed here under the terms of Article 25fa of the Dutch Copyright Act, indicated by the “Taverne” license. -

Ribosomal Proteins and Human Diseases: Molecular Mechanisms and Targeted Therapy ✉ Jian Kang1,2, Natalie Brajanovski1, Keefe T
Signal Transduction and Targeted Therapy www.nature.com/sigtrans REVIEW ARTICLE OPEN Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy ✉ Jian Kang1,2, Natalie Brajanovski1, Keefe T. Chan1,2, Jiachen Xuan1,2, Richard B. Pearson1,2,3,4 and Elaine Sanij1,2,5,6 Ribosome biogenesis and protein synthesis are fundamental rate-limiting steps for cell growth and proliferation. The ribosomal proteins (RPs), comprising the structural parts of the ribosome, are essential for ribosome assembly and function. In addition to their canonical ribosomal functions, multiple RPs have extra-ribosomal functions including activation of p53-dependent or p53- independent pathways in response to stress, resulting in cell cycle arrest and apoptosis. Defects in ribosome biogenesis, translation, and the functions of individual RPs, including mutations in RPs have been linked to a diverse range of human congenital disorders termed ribosomopathies. Ribosomopathies are characterized by tissue-specific phenotypic abnormalities and higher cancer risk later in life. Recent discoveries of somatic mutations in RPs in multiple tumor types reinforce the connections between ribosomal defects and cancer. In this article, we review the most recent advances in understanding the molecular consequences of RP mutations and ribosomal defects in ribosomopathies and cancer. We particularly discuss the molecular basis of the transition from hypo- to hyper-proliferation in ribosomopathies with elevated cancer risk, a paradox termed “Dameshek’s riddle.” Furthermore, we review the current treatments for ribosomopathies and prospective therapies targeting ribosomal defects. We also highlight recent advances in ribosome stress-based cancer therapeutics. Importantly, insights into the mechanisms of resistance to therapies targeting ribosome biogenesis bring new perspectives into the molecular basis of cancer susceptibility in ribosomopathies and new 1234567890();,: clinical implications for cancer therapy. -

Vast Human-Specific Delay in Cortical Ontogenesis Associated With
Supplementary information Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques Supplementary Methods Sample collection We used prefrontal cortex (PFC) and cerebellar cortex (CBC) samples from postmortem brains of 33 human (aged 0-98 years), 14 chimpanzee (aged 0-44 years) and 44 rhesus macaque individuals (aged 0-28 years) (Table S1). Human samples were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, USA, the Netherlands Brain Bank, Amsterdam, Netherlands and the Chinese Brain Bank Center, Wuhan, China. Informed consent for use of human tissues for research was obtained in writing from all donors or their next of kin. All subjects were defined as normal by forensic pathologists at the corresponding brain bank. All subjects suffered sudden death with no prolonged agonal state. Chimpanzee samples were obtained from the Yerkes Primate Center, GA, USA, the Anthropological Institute & Museum of the University of Zürich-Irchel, Switzerland and the Biomedical Primate Research Centre, Netherlands (eight Western chimpanzees, one Central/Eastern and five of unknown origin). Rhesus macaque samples were obtained from the Suzhou Experimental Animal Center, China. All non-human primates used in this study suffered sudden deaths for reasons other than their participation in this study and without any relation to the tissue used. CBC dissections were made from the cerebellar cortex. PFC dissections were made from the frontal part of the superior frontal gyrus. All samples contained an approximately 2:1 grey matter to white matter volume ratio. RNA microarray hybridization RNA isolation, hybridization to microarrays, and data preprocessing were performed as described previously (Khaitovich et al.