Structural Studies on a Ribonucleoprotein Organelle: the Ribosome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Size Changes in Eukaryotic Ribosomes
Proc. Nat. Acad. Sci. USA Vol. 68, No. 12, pp. 3021-3025, December 1971 Size Changes in Eukaryotic Ribosomes (diffusion constant/sedimentation constant/ribosomal dissociation/chick embryo) JOHN VOURNAKIS AND ALEXANDER RICH Department of Biology, Massachusetts Institute of Technology, Cambridge, Mass. 02139 Contributed by Alexander Rich, September 20, 1971 ABSTRACT Evidence is presented that ribosomes two particles are similar. However, these changes suggest active in protein synthesis and attached to messenger that when the ribosome is attached to messenger RNA it has a RNA on polysomes have a smaller diameter than free cytoplasmic single ribosomes. Measurements have been smaller diameter than is found for the free cytoplastic ribo- made on these two types of ribosomes of differences in some. This more compact form of the ribosome is maintained sedimentation velocity and diffusion constant. Differences even when the nascent polypeptide chain is relased by puro- in these quantities suggest about a 20-A decrease in the mycin. We thus infer that there are substantial differences diameter of the ribosomes from chick embryo muscles in the interactions between the ribosomal subunits when when they are attached to messenger RNA. Similar dif- they ferences are also observed in rabbit reticulocytes and are attached to messenger RNA as compared to the free cyto- mouse ascites tumor cells. These two ribosomal states plasmic single ribosome, which is inactive in protein synthesis. have different sensitivity to Pronase digestion and dis- sociate into ribosomal subunits at different KCI concen- METHODS AND MATERIALS trations. This size difference is not associated with a sig- nificant difference in overall ribosomal mass and appears Preparation of Ribosomes. -

A Global Analysis of Enzyme Compartmentalization to Glycosomes
pathogens Article A Global Analysis of Enzyme Compartmentalization to Glycosomes Hina Durrani 1, Marshall Hampton 2 , Jon N. Rumbley 3 and Sara L. Zimmer 1,* 1 Department of Biomedical Sciences, University of Minnesota Medical School, Duluth Campus, Duluth, MN 55812, USA; [email protected] 2 Mathematics & Statistics Department, University of Minnesota Duluth, Duluth, MN 55812, USA; [email protected] 3 College of Pharmacy, University of Minnesota, Duluth Campus, Duluth, MN 55812, USA; [email protected] * Correspondence: [email protected] Received: 25 March 2020; Accepted: 9 April 2020; Published: 12 April 2020 Abstract: In kinetoplastids, the first seven steps of glycolysis are compartmentalized into a glycosome along with parts of other metabolic pathways. This organelle shares a common ancestor with the better-understood eukaryotic peroxisome. Much of our understanding of the emergence, evolution, and maintenance of glycosomes is limited to explorations of the dixenous parasites, including the enzymatic contents of the organelle. Our objective was to determine the extent that we could leverage existing studies in model kinetoplastids to determine the composition of glycosomes in species lacking evidence of experimental localization. These include diverse monoxenous species and dixenous species with very different hosts. For many of these, genome or transcriptome sequences are available. Our approach initiated with a meta-analysis of existing studies to generate a subset of enzymes with highest evidence of glycosome localization. From this dataset we extracted the best possible glycosome signal peptide identification scheme for in silico identification of glycosomal proteins from any kinetoplastid species. Validation suggested that a high glycosome localization score from our algorithm would be indicative of a glycosomal protein. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Cilia and Flagella: from Discovery to Disease Dylan J
Dartmouth Undergraduate Journal of Science Volume 20 Article 2 Number 1 Assembly 2017 Cilia and Flagella: From Discovery to Disease Dylan J. Cahill Dylan Cahill, [email protected] Follow this and additional works at: https://digitalcommons.dartmouth.edu/dujs Part of the Engineering Commons, Life Sciences Commons, Medicine and Health Sciences Commons, Physical Sciences and Mathematics Commons, and the Social and Behavioral Sciences Commons Recommended Citation Cahill, Dylan J. (2017) "Cilia and Flagella: From Discovery to Disease," Dartmouth Undergraduate Journal of Science: Vol. 20 : No. 1 , Article 2. Available at: https://digitalcommons.dartmouth.edu/dujs/vol20/iss1/2 This Research Article is brought to you for free and open access by the Student-led Journals and Magazines at Dartmouth Digital Commons. It has been accepted for inclusion in Dartmouth Undergraduate Journal of Science by an authorized editor of Dartmouth Digital Commons. For more information, please contact [email protected]. BIOLOGY Cilia and Flagella: FromCilia and Discovery Flagella: to Disease From Discovery to Disease BY DYLAN CAHILL ‘18 Introduction certain insect sperm fagella (3, 5, 6). A unique Figure 1: Chlamydomonas intracellular transport mechanism known as reinhardtii, a single-celled, bi- In 1674, peering through the lens of a crude flagellate green alga, viewed intrafagellar transport is responsible for the light microscope, Antoni van Leeuwenhoek with a scanning electron assembly and maintenance of these organelles Chlamydomonas observed individual living cells for the frst time microscope. is (3, 6). Cilia and fagella are primarily composed a model organism in flagellar in history (1). He noted long, thin appendages of the protein tubulin, which polymerizes into dynamics and motility studies. -

Organization and Evolution of 5S Ribosomal Dna in the Fish Genome
In: Focus on Genome Research ISSN 1-59033-960-6 Editor: Clyde R. Williams, pp335-363 ©2004 Nova Science Publishers, Inc. Chapter X ORGANIZATION AND EVOLUTION OF 5S RIBOSOMAL DNA IN THE FISH GENOME Cesar Martins 1 and Adriane Pinto Wasko 2 Departamento de Morfologia, Instituto de Biociências, Universidade Estadual Paulista, CEP 18618-000, Botucatu, SP, Brazil. Phone/Fax +55 14 38116264, 1e-mail [email protected]; 2e-mail: [email protected] ABSTRACT In higher eukaryotes, the 5S ribosomal multigene family (5S rDNA) is tandemly organized in repeat units composed of a coding region (5S rRNA gene) and a non-transcribed spacer sequence (NTS). Although the 5S rDNA organization has been investigated in several vertebrate species, present data are concentrated in mammals and amphibians, whereas other groups, such as fishes, have been poorly studied. To further the understanding on the dynamics and evolution of 5S rDNA arrays in the vertebrate genome, recent studies have focused on the genome organization of these sequences in fish species, which represent the base group of vertebrate evolution. It was evidenced that the chromosome distribution of the 5S rDNA is quite conserved among related fish species occupying an interstitial position in the chromosomes. Although the 5S rDNA clusters have been maintained conserved in the chromosomes, changes in the nucleotide sequences and organization of the repeat units have occurred in fish species, as demonstrated by the presence of 5S rDNA variant types within and between genomes clustered in distinct chromosome environments. These variants are distributed in two major classes, suggesting that such pattern could represent a primitive condition for the fish genome, as well as for vertebrates. -

The Flagellum and Flagellar Pocket of Trypanosomatids
Molecular & Biochemical Parasitology 115 (2001) 1–17 www.parasitology-online.com. Reviews: Parasite cell Biology: 1 The flagellum and flagellar pocket of trypanosomatids Scott M. Landfear *, Marina Ignatushchenko Department of Molecular Microbiology and Immunology, Oregon Health Sciences Uni6ersity, Portland, OR 97201, USA Received 9 November 2000; received in revised form 26 January 2001; accepted 5 March 2001 Abstract The flagellum and flagellar pocket are distinctive organelles present among all of the trypanosomatid protozoa. Currently, recognized functions for these organelles include generation of motility for the flagellum and dedicated secretory and endocytic activities for the flagellar pocket. The flagellar and flagellar pocket membranes have long been recognized as morphologically separate domains that are component parts of the plasma membrane that surrounds the entire cell. The structural and functional specialization of these two membranes has now been underscored by the identification of multiple proteins that are targeted selectively to each of these domains, and non-membrane proteins have also been identified that are targeted to the internal lumina of these organelles. Investigations on the functions of these organelle-specific proteins should continue to shed light on the unique biological activities of the flagellum and flagellar pocket. In addition, work has begun on identifying signals or modifications of these proteins that direct their targeting to the correct subcellular location. Future endeavors should further refine our knowledge of targeting signals and begin to dissect the molecular machinery involved in transporting and retaining each polypeptide at its designated cellular address. © 2001 Elsevier Science B.V. All rights reserved. Keywords: Trypanosomatid protozoa; Flagellum; Flagellar Pocket; Organelle-specific proteins; Review 1. -
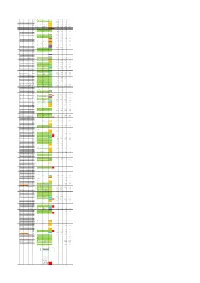
End Strand of Gene Gene Name Gene Function Starnd of Trascript
Genomic TSS strand Gene starnd of Transcipt relatively to TSS Start End Gene name coordinates relatively TSS type Control_fwd TEX_fwd Control_rev TEX_rev of gene function trascript gene group of TSS to ATG ribosomal 93 854 + rps12 + 1 protein S12 1 1 14.0638317 20.7539794 0 0 93 324 exon 1 + 111 rps12 intron 1 1 13.90756687 18.07149224 0.781323982 0.423550599 829 854 exon 2 + 496 rps12 exon2 2 24.22104343 15.24782157 0 0 30S 904 1371 + gene rps7 ribosomal + 2 protein S7 1303 -209 rps7 inter (ndhB) 2 20.47068832 9.035746118 0.625059185 0.847101199 NADH dehydrogen 1512 3535 + gene ndhB (ndh2) + ase subunit 3 2 1696 ndhB exon 1-inter 2 3.594090315 2.964854195 0.468794389 0.282367066 + 2209 ndhB exon 1-inter 2 46.09811492 4.09432246 0.468794389 0.423550599 1512 2237 exon 1 + 2756 ndhB exon 2 -inter 2 43.28534858 4.800240125 0.312529593 0.282367066 2756 3535 exon 2 + 3090 ndhB exon 2 -inter 2 17.50165719 15.95373924 0.312529593 0 + 3192 ndhB exon 2 -inter 2 140.6383167 117.6058831 2.812766334 1.694202397 - 3462 ndhB exon 2 -inter 3 1.406383167 1.129468265 1.406383167 3.67077186 4 3633 3712 + tRNA tRNA-Leu-CAA + 3610 -23 tRNA-Leu 1 77.19480938 84.85130339 0.625059185 0 + 3633 1 tRNA-Leu 1 359.5652963 649.0207016 0.781323982 0 photosyste 3954 4058 + gene psbM m II protein + 5 M 3775 -179 psbM 2 20.47068832 12.00060031 0 0.141183533 + 3954 psbM-0 2 69.22530477 28.37789015 0.156264796 0 hypothetical 4182 5073 + gene ycf66 6 protein 4182 4287 exon 1 4772 5073 exon 2 7 5202 5113 - gene ycf (ORF29) - 5299 orf29 inter 1 0 0 3.125295926 3.67077186 -

HUMAN RIBOSOME BIOGENESIS and the REGULATION of the TUMOUR SUPPRESSOR P53
HUMAN RIBOSOME BIOGENESIS AND THE REGULATION OF THE TUMOUR SUPPRESSOR p53 Andria Pelava Submitted for Doctor of Philosophy Final submission: December 2016 Institute of Cell and Molecular Biosciences Faculty of Medical Sciences Newcastle University ii Abstract Ribosome production is an energetically expensive and, therefore, highly regulated process. Defects in ribosome biogenesis lead to genetic diseases called Ribosomopathies, such as Dyskeratosis Congenita (DC), and mutations in ribosomal proteins and ribosome biogenesis factors are linked to multiple types of cancer. During ribosome biogenesis, the ribosomal RNAs (rRNAs) are processed and modified, and defects in ribosome biogenesis lead to the activation of p53. This project aimed to investigate the functions of Dyskerin, mutated in X-linked DC, in human ribosome biogenesis and p53 regulation, and to explore the link between ribosome production and p53 homeostasis. Dyskerin is an rRNA pseudouridine synthase and required for telomere maintenance. There is some debate as to whether DC is the result of telomere maintenance or ribosome biogenesis defects. It is shown here that human Dyskerin is required for the production of both LSU and SSU, and knockdown of Dyskerin leads to p53 activation via inhibition of MDM2 via the 5S RNP, an LSU assembly intermediate which accumulates after ribosome biogenesis defects. My data indicate that p53 activation, due to defects in ribosome biogenesis, may contribute to the clinical symptoms seen in patients suffering with DC. In addition, it is shown that defects in early or late stages of SSU or LSU biogenesis, result in activation of p53 via the 5S RNP-MDM2 pathway, and that p53 is induced in less than 12 hours after ribosome biogenesis defects. -

Selenoprotein H Is an Essential Regulator of Redox Homeostasis That
Selenoprotein H is an essential regulator of redox PNAS PLUS homeostasis that cooperates with p53 in development and tumorigenesis Andrew G. Coxa, Allison Tsomidesa, Andrew J. Kima, Diane Saundersa, Katie L. Hwanga, Kimberley J. Evasonb, Jerry Heidelc, Kristin K. Brownd, Min Yuand, Evan C. Liend, Byung Cheon Leea,e, Sahar Nissima, Bryan Dickinsonf, Sagar Chhangawalag, Christopher J. Changh,i, John M. Asarad, Yariv Houvrasg, Vadim N. Gladysheva,j, and Wolfram Goesslinga,j,k,l,1 aBrigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115; bUniversity of Utah, Salt Lake City, UT 84112; cOregon State University, Corvallis, OR 97331; dBeth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02115; eKorea University, 02841 Seoul, Republic of Korea; fUniversity of Chicago, Chicago, IL 60637; gWeill Cornell Medical College and New York Presbyterian Hospital, New York, NY 10065; hHoward Hughes Medical Institute, Bethesda, MD 20815; iUniversity of California, Berkeley, CA 20815; jBroad Institute of MIT and Harvard, Cambridge, MA 02142; kHarvard Stem Cell Institute, Cambridge, MA 02138; and lDana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115 Edited by Leonard I. Zon, Howard Hughes Medical Institute, Boston Children’s Hospital, Harvard Medical School, Boston, MA, and accepted by Editorial Board Member Carol Prives July 18, 2016 (received for review January 7, 2016) Selenium, an essential micronutrient known for its cancer preven- mice exhibit impaired selenium transport from the liver to pe- tion properties, is incorporated into a class of selenocysteine- ripheral tissues and show growth retardation and impaired motor containing proteins (selenoproteins). Selenoprotein H (SepH) is a coordination (13, 14). -

Driving Nucleolar Assembly
Downloaded from genesdev.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press PERSPECTIVE Driving nucleolar assembly Kathleen L. McCann,1 and Susan J. Baserga1,2,3,4 1Department of Genetics, 2Department of Molecular Biophysics and Biochemistry, 3Department of Therapeutic Radiology, Yale University School of Medicine, New Haven, Connecticut 06520, USA In this issue of Genes & Development,Grobandcol- breaks down (open mitosis), transcription of the ribosomal leagues (pp. 220–230) identify the minimal molecular re- RNA (rRNA) is inhibited, and the nucleolus disassem- quirements to assemble a fully functional nucleolus in bles. Upon completion of mitosis, rRNA transcription human cells and demonstrate the importance of the is reinitiated within the NOR, ribosome biogenesis fac- nucleolar transcription factor upstream binding factor tors are recruited, and the nucleolus is assembled. Only (UBF) as a mitotic bookmark at the ribosomal DNA NORs that are actively engaged in transcription can (rDNA). direct nucleolar assembly (Hernandez-Verdun 2011). Consequently, the nucleolus truly appears to be ‘‘an organelle formed by the act of building a ribosome’’ (Melese and Xue 1995). In all eukaryotes, the nucleolus is an essential, non- To begin to reveal the molecular mechanisms of nucle- membrane-bound organelle within the nucleus. Assem- olar formation, McStay’s laboratory (Mais et al. 2005; Grob bled around the ribosomal DNA (rDNA), the nucleolus et al. 2014) has applied synthetic biology. In an earlier is the site of ribosome biogenesis. In addition to its role report, they introduced 6.4 kb of DNA repeat sequences in making ribosomes, the nucleolus functions in other from the intergenic spacer of the Xenopus ribosomal gene important cellular processes, including stress sensing, into a noncanonical site in the human genome. -

UC Riverside Electronic Theses and Dissertations
UC Riverside UC Riverside Electronic Theses and Dissertations Title Characterization of AtRAP Function in Plant Immunity and in RNA Transportation Permalink https://escholarship.org/uc/item/8pk9k169 Author Wang, Huan Publication Date 2017 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA RIVERSIDE Characterization of AtRAP Function in Plant Immunity and in RNA Transportation A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Plant Pathology by Huan Wang December 2017 Dissertation Committee: Dr. Hailing Jin, Chairperson Dr. Julia Bailey-Serres Dr. Isgouhi Kaloshian Copyright by Huan Wang 2017 The Dissertation of Huan Wang is approved: Committee Chairperson University of California, Riverside ACKNOWLEDGEMENTS I would like to express my sincere gratitude and appreciation to all those who helped and encouraged me during my graduate study. My deepest gratitude goes first and foremost to Dr. Hailing Jin, my supervisor, for her patient and constant guidance and encouragement. No matter what the difficulty is, she always stands with me to give me very timely, experienced and smart advice to help me solve it quickly and efficiently. Her creative ideas, intelligent analysis, hardworking attitude and generous personality inspire and teach me a lot not only on my research but also on my philosophy of life. Secondly, I would like to express my heartfelt gratitude to my dissertation committee members, Dr. Julia Bailey-Serres and Dr. Isgouhi Kaloshian. I really appreciate your valuable suggestions and kindly help to my study. Dr. Julia Bailey-Serres continuously helps my study to be my guidance committee member, qualifying exam committee member, and dissertation committee member. -

Plastid in Human Parasites
SCIENTIFIC CORRESPONDENCE being otherwise homo Plastid in human geneous. The 35-kb genome-containing organ parasites elle identified here did not escape the attention of early Sm - The discovery in malarial and toxo electron microscopists who plasmodial parasites of genes normally - not expecting the pres occurring in the photosynthetic organelle ence of a plastid in a proto of plants and algae has prompted specula zoan parasite like tion that these protozoans might harbour Toxoplasma - ascribed to it a vestigial plastid1• The plastid-like para various names, including site genes occur on an extrachromosomal, 'Hohlzylinder' (hollow cylin maternally inherited2, 35-kilobase DNA der), 'Golgi adjunct' and circle with an architecture reminiscent of 'grof3e Tilkuole mit kriiftiger that of plastid genomes3•4• Although the Wandung' (large vacuole 35-kb genome is distinct from the 6-7-kb with stout surrounds) ( see linear mitochondrial genome3-6, it is not refs cited in ref. 9). Our pre known where in the parasite cells the plas liminary experiments with tid-like genome resides. Plasmodium falciparum, the To determine whether a plastid is pre causative agent of the most sent, we used high-resolution in situ lethal form of malaria, hybridization7 to localize transcripts of a identify an organelle (not plastid-like 16S ribosomal RNA gene shown) which appears sim from Toxoplasma gondii8, the causative ilar to the T. gondii plastid. agent of toxoplasmosis. Transcripts accu The number of surrounding mulate in a small, ovoid organelle located membranes in the P. anterior to the nucleus in the mid-region falciparum plastid, and its of the cell (a, b in the figure).