Driving Nucleolar Assembly
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Size Changes in Eukaryotic Ribosomes
Proc. Nat. Acad. Sci. USA Vol. 68, No. 12, pp. 3021-3025, December 1971 Size Changes in Eukaryotic Ribosomes (diffusion constant/sedimentation constant/ribosomal dissociation/chick embryo) JOHN VOURNAKIS AND ALEXANDER RICH Department of Biology, Massachusetts Institute of Technology, Cambridge, Mass. 02139 Contributed by Alexander Rich, September 20, 1971 ABSTRACT Evidence is presented that ribosomes two particles are similar. However, these changes suggest active in protein synthesis and attached to messenger that when the ribosome is attached to messenger RNA it has a RNA on polysomes have a smaller diameter than free cytoplasmic single ribosomes. Measurements have been smaller diameter than is found for the free cytoplastic ribo- made on these two types of ribosomes of differences in some. This more compact form of the ribosome is maintained sedimentation velocity and diffusion constant. Differences even when the nascent polypeptide chain is relased by puro- in these quantities suggest about a 20-A decrease in the mycin. We thus infer that there are substantial differences diameter of the ribosomes from chick embryo muscles in the interactions between the ribosomal subunits when when they are attached to messenger RNA. Similar dif- they ferences are also observed in rabbit reticulocytes and are attached to messenger RNA as compared to the free cyto- mouse ascites tumor cells. These two ribosomal states plasmic single ribosome, which is inactive in protein synthesis. have different sensitivity to Pronase digestion and dis- sociate into ribosomal subunits at different KCI concen- METHODS AND MATERIALS trations. This size difference is not associated with a sig- nificant difference in overall ribosomal mass and appears Preparation of Ribosomes. -

The Splicing Factor XAB2 Interacts with ERCC1-XPF and XPG for RNA-Loop Processing During Mammalian Development
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.20.211441; this version posted July 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The Splicing Factor XAB2 interacts with ERCC1-XPF and XPG for RNA-loop processing during mammalian development Evi Goulielmaki1*, Maria Tsekrekou1,2*, Nikos Batsiotos1,2, Mariana Ascensão-Ferreira3, Eleftheria Ledaki1, Kalliopi Stratigi1, Georgia Chatzinikolaou1, Pantelis Topalis1, Theodore Kosteas1, Janine Altmüller4, Jeroen A. Demmers5, Nuno L. Barbosa-Morais3, George A. Garinis1,2* 1. Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology- Hellas, GR70013, Heraklion, Crete, Greece, 2. Department of Biology, University of Crete, Heraklion, Crete, Greece, 3. Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa, Avenida Professor Egas Moniz, 1649-028 Lisboa, Portugal, 4. Cologne Center for Genomics (CCG), Institute for Genetics, University of Cologne, 50931, Cologne, Germany, 5. Proteomics Center, Netherlands Proteomics Center, and Department of Biochemistry, Erasmus University Medical Center, the Netherlands. Corresponding author: George A. Garinis ([email protected]) *: equally contributing authors bioRxiv preprint doi: https://doi.org/10.1101/2020.07.20.211441; this version posted July 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract RNA splicing, transcription and the DNA damage response are intriguingly linked in mammals but the underlying mechanisms remain poorly understood. Using an in vivo biotinylation tagging approach in mice, we show that the splicing factor XAB2 interacts with the core spliceosome and that it binds to spliceosomal U4 and U6 snRNAs and pre-mRNAs in developing livers. -

A Global Analysis of Enzyme Compartmentalization to Glycosomes
pathogens Article A Global Analysis of Enzyme Compartmentalization to Glycosomes Hina Durrani 1, Marshall Hampton 2 , Jon N. Rumbley 3 and Sara L. Zimmer 1,* 1 Department of Biomedical Sciences, University of Minnesota Medical School, Duluth Campus, Duluth, MN 55812, USA; [email protected] 2 Mathematics & Statistics Department, University of Minnesota Duluth, Duluth, MN 55812, USA; [email protected] 3 College of Pharmacy, University of Minnesota, Duluth Campus, Duluth, MN 55812, USA; [email protected] * Correspondence: [email protected] Received: 25 March 2020; Accepted: 9 April 2020; Published: 12 April 2020 Abstract: In kinetoplastids, the first seven steps of glycolysis are compartmentalized into a glycosome along with parts of other metabolic pathways. This organelle shares a common ancestor with the better-understood eukaryotic peroxisome. Much of our understanding of the emergence, evolution, and maintenance of glycosomes is limited to explorations of the dixenous parasites, including the enzymatic contents of the organelle. Our objective was to determine the extent that we could leverage existing studies in model kinetoplastids to determine the composition of glycosomes in species lacking evidence of experimental localization. These include diverse monoxenous species and dixenous species with very different hosts. For many of these, genome or transcriptome sequences are available. Our approach initiated with a meta-analysis of existing studies to generate a subset of enzymes with highest evidence of glycosome localization. From this dataset we extracted the best possible glycosome signal peptide identification scheme for in silico identification of glycosomal proteins from any kinetoplastid species. Validation suggested that a high glycosome localization score from our algorithm would be indicative of a glycosomal protein. -

Nucleolus: a Central Hub for Nuclear Functions Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, Sergey Razin, Yegor Vassetzky
Nucleolus: A Central Hub for Nuclear Functions Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, Sergey Razin, Yegor Vassetzky To cite this version: Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, et al.. Nucleolus: A Central Hub for Nuclear Functions. Trends in Cell Biology, Elsevier, 2019, 29 (8), pp.647-659. 10.1016/j.tcb.2019.04.003. hal-02322927 HAL Id: hal-02322927 https://hal.archives-ouvertes.fr/hal-02322927 Submitted on 18 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Nucleolus: A Central Hub for Nuclear Functions Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, Sergey Razin, Yegor Vassetzky To cite this version: Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, et al.. Nucleolus: A Central Hub for Nuclear Functions. Trends in Cell Biology, Elsevier, 2019, 29 (8), pp.647-659. 10.1016/j.tcb.2019.04.003. hal-02322927 HAL Id: hal-02322927 https://hal.archives-ouvertes.fr/hal-02322927 Submitted on 18 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. -

Unit 6-Nucleus
Unit 6-Nucleus Dr. Pallee shree Nucleus • Nucleus is the most important organelle in the cell • It distinguishes eukaryotic from prokaryotic cells • By housing the cell's genome, the nucleus serves both as the repository of genetic information and as the cell's control center • DNA replication, transcription, and RNA processing all take place within the nucleus Cont… • A nucleus is a double-membraned eukaryotic cell organelle that contains the genetic material. • It appears in an oval shape averages 5µm in width. • It often lies in the centre of a cell • The nucleus was the first organelle to be discovered • Nuclei 1st discovered and named by Robert Brown • Role of nucleus 1st demonestrated by Max Hammerling Ultra structure of Nucleus 1. Nuclear envelope 2. nuclear pores 3. Nucleoplasm 4. Nucleolus 5. Chromosomes 1. Structure of Nuclear envelope • The nuclear envelope has a complex structure consisting of a) Two nuclear membranes separated by a perinuclear space measuring about 20–40 nm across b) Underlying nuclear lamina • The nucleus is surrounded by a system of two concentric membranes, called the inner and outer nuclear membranes • The inner and outer nuclear membranes are joined at nuclear pore complexes a. Nuclear membranes • The outer nuclear membrane is continuous with the endoplasmic reticulum, so the space between the • The critical function of the inner and outer nuclear membranes nuclear membranes is to act as is directly connected with the lumen a barrier that separates the of the ER contents of the nucleus from the cytoplasm. • It is functionally similar to the membranes of the ER and has • Like other cell membranes, ribosomes bound to its cytoplasmic each nuclear membrane is a surface but protein composition phospholipid bilayer differ slightly as they are enriched in permeable only to small proteins which binds to cytoskeleton nonpolar molecules • The inner nuclear membrane carries • Other molecules are unable to proteins that are specific to the diffuse through the bilayer. -

Cilia and Flagella: from Discovery to Disease Dylan J
Dartmouth Undergraduate Journal of Science Volume 20 Article 2 Number 1 Assembly 2017 Cilia and Flagella: From Discovery to Disease Dylan J. Cahill Dylan Cahill, [email protected] Follow this and additional works at: https://digitalcommons.dartmouth.edu/dujs Part of the Engineering Commons, Life Sciences Commons, Medicine and Health Sciences Commons, Physical Sciences and Mathematics Commons, and the Social and Behavioral Sciences Commons Recommended Citation Cahill, Dylan J. (2017) "Cilia and Flagella: From Discovery to Disease," Dartmouth Undergraduate Journal of Science: Vol. 20 : No. 1 , Article 2. Available at: https://digitalcommons.dartmouth.edu/dujs/vol20/iss1/2 This Research Article is brought to you for free and open access by the Student-led Journals and Magazines at Dartmouth Digital Commons. It has been accepted for inclusion in Dartmouth Undergraduate Journal of Science by an authorized editor of Dartmouth Digital Commons. For more information, please contact [email protected]. BIOLOGY Cilia and Flagella: FromCilia and Discovery Flagella: to Disease From Discovery to Disease BY DYLAN CAHILL ‘18 Introduction certain insect sperm fagella (3, 5, 6). A unique Figure 1: Chlamydomonas intracellular transport mechanism known as reinhardtii, a single-celled, bi- In 1674, peering through the lens of a crude flagellate green alga, viewed intrafagellar transport is responsible for the light microscope, Antoni van Leeuwenhoek with a scanning electron assembly and maintenance of these organelles Chlamydomonas observed individual living cells for the frst time microscope. is (3, 6). Cilia and fagella are primarily composed a model organism in flagellar in history (1). He noted long, thin appendages of the protein tubulin, which polymerizes into dynamics and motility studies. -

The Flagellum and Flagellar Pocket of Trypanosomatids
Molecular & Biochemical Parasitology 115 (2001) 1–17 www.parasitology-online.com. Reviews: Parasite cell Biology: 1 The flagellum and flagellar pocket of trypanosomatids Scott M. Landfear *, Marina Ignatushchenko Department of Molecular Microbiology and Immunology, Oregon Health Sciences Uni6ersity, Portland, OR 97201, USA Received 9 November 2000; received in revised form 26 January 2001; accepted 5 March 2001 Abstract The flagellum and flagellar pocket are distinctive organelles present among all of the trypanosomatid protozoa. Currently, recognized functions for these organelles include generation of motility for the flagellum and dedicated secretory and endocytic activities for the flagellar pocket. The flagellar and flagellar pocket membranes have long been recognized as morphologically separate domains that are component parts of the plasma membrane that surrounds the entire cell. The structural and functional specialization of these two membranes has now been underscored by the identification of multiple proteins that are targeted selectively to each of these domains, and non-membrane proteins have also been identified that are targeted to the internal lumina of these organelles. Investigations on the functions of these organelle-specific proteins should continue to shed light on the unique biological activities of the flagellum and flagellar pocket. In addition, work has begun on identifying signals or modifications of these proteins that direct their targeting to the correct subcellular location. Future endeavors should further refine our knowledge of targeting signals and begin to dissect the molecular machinery involved in transporting and retaining each polypeptide at its designated cellular address. © 2001 Elsevier Science B.V. All rights reserved. Keywords: Trypanosomatid protozoa; Flagellum; Flagellar Pocket; Organelle-specific proteins; Review 1. -

Functional Proximity Mapping of RNA Binding Proteins Uncovers a Mitochondrial Mrna Anchor That Promotes Stress Recovery
Functional proximity mapping of RNA binding proteins uncovers a mitochondrial mRNA anchor that promotes stress recovery Wei Qin Stanford University Samuel Myers Broad Institute Dominique Carey The Broad Institute of MIT and Harvard Steven Carr Broad Institute Alice Ting ( [email protected] ) Stanford University https://orcid.org/0000-0002-8277-5226 Article Keywords: Unbiased Proteome Discovery, Nucleolus, Outer Mitochondrial Membrane, Local Translation, Functional Protein Mapping Posted Date: November 12th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-103196/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/49 Abstract Proximity labeling (PL) with genetically-targeted promiscuous enzymes has emerged as a powerful tool for unbiased proteome discovery. By combining the spatiotemporal specicity of PL with methods for functional protein enrichment, it should be possible to map specic protein subclasses within distinct compartments of living cells. Here we demonstrate this capability for RNA binding proteins (RBPs), by combining peroxidase-based PL with organic-aqueous phase separation of crosslinked protein-RNA complexes (“APEX-PS”). We validated APEX-PS by mapping nuclear RBPs, then applied it to uncover the RBPomes of two unpuriable subcompartments - the nucleolus and the outer mitochondrial membrane (OMM). At the OMM, we discovered the RBP SYNJ2BP, which retains specic nuclear-encoded mitochondrial mRNAs during translation stress, to promote their local translation and import of protein products into the mitochondrion during stress recovery. APEX-PS is a versatile tool for compartment- specic RBP discovery and expands the scope of PL to functional protein mapping. Introduction RNA-protein interactions are pervasive in both transient and stable macromolecular complexes underlying transcription, translation and stress response 1, 2. -
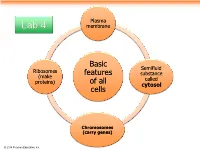
Basic Features of All Cells
Plasma Lab 4 membrane Basic Semifluid Ribosomes features substance (make called proteins) of all cytosol cells Chromosomes (carry genes) © 2014 Pearson Education, Inc. Prokaryotic cells are characterized by having . No nucleus . DNA in an unbound region called the nucleoid . No membrane-bound organelles . Cytoplasm bound by the plasma membrane © 2014 Pearson Education, Inc. Eukaryotic cells are characterized by having • DNA in a nucleus that is bounded by a membranous nuclear envelope • Membrane-bound organelles • Cytoplasm in the region between the plasma membrane and nucleus Eukaryotic cells are generally much larger than prokaryotic cells © 2014 Pearson Education, Inc. Figure 6.8a ENDOPLASMIC RETICULUM (ER) Nuclear envelope Rough ER Smooth ER Nucleolus NUCLEUS Flagellum Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes Microvilli Golgi apparatus Peroxisome Lysosome Mitochondrion © 2014 Pearson Education, Inc. Figure 6.8b Nuclear envelope NUCLEUS Nucleolus Rough ER Chromatin Smooth ER Ribosomes Golgi Central vacuole apparatus Microfilaments CYTOSKELETON Microtubules Mitochondrion Peroxisome Plasma Chloroplast membrane Cell wall Plasmodesmata Wall of adjacent cell © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. Table 6.1 © 2014 Pearson Education, Inc. Cell Walls of Plants The cell wall is an extracellular structure that distinguishes plant cells from animal cells Plant cell walls Prokaryotes, are made -

Segregation of the Nucleolus Produced by Anthramycin1
[CANCER RESEARCH 28, 81-90, January 1968] Segregation of the Nucleolus Produced by Anthramycin1 Curtis Harris, Harold Grady, and Donald Svoboda Department oj Pathology, University oj Kansas Medical Center, Kansas City, Kansas 66103 SUMMARY mycin does not produce toxicity in the bone marrow and gastrointestinal tract (49). It possesses certain structural fea Anthramycin, an antibiotic possessing antitumor properties, tures in common with actinomycin D and the postulated biosyn- was administered to rats and mice in a single intraperitoneal thetic intermediates of the actinomycin^, 3-hydroxy-4-methyI- close ranging from 1 to 10 mg/kg body weight. Light micro anthraniloyl peptides (3,47). Like kanamycin, neomycin, and scopy showed that, at the highest dose, anthramycin produced other antibiotics (48), anthramycin permanently bleaches necrosis in the pancreas of rats but not in mice. By electron Euglena gracilis (11). microscopy, nucleolar changes were observed in the spleen and This report demonstrates that anthramycin is another agent kidney of both species by 1 hour. Acinar cells of the pancreas which causes nucleolar changes in pancreatic cells, reticulo showed nucleolar caps and segregation of nucleolar constituents endothelial cells of liver and spleen, and in proximal tubular in both species at 3 to 4 hours. By 10 hours segregation of the epithelium in the kidney. nucleolus and focal cytoplasmic degradation were prominent in pancreatic cells of rats given higher doses but were not MATERIALS AND METHODS observed in hepatocytes of either species. Anthramycin produced nucleolar alterations in Kupffer cells, Thirty-six inbred Fisher-344 male rats weighing between 95 endocrine and exocrine pancreatic cells, proximal tubular cells and 240 gm and fifteen C57 black mice weighing between 20 of the kidney, and in the reticuloendothelial cells of the spleen and 30 gm were used. -

Plastid in Human Parasites
SCIENTIFIC CORRESPONDENCE being otherwise homo Plastid in human geneous. The 35-kb genome-containing organ parasites elle identified here did not escape the attention of early Sm - The discovery in malarial and toxo electron microscopists who plasmodial parasites of genes normally - not expecting the pres occurring in the photosynthetic organelle ence of a plastid in a proto of plants and algae has prompted specula zoan parasite like tion that these protozoans might harbour Toxoplasma - ascribed to it a vestigial plastid1• The plastid-like para various names, including site genes occur on an extrachromosomal, 'Hohlzylinder' (hollow cylin maternally inherited2, 35-kilobase DNA der), 'Golgi adjunct' and circle with an architecture reminiscent of 'grof3e Tilkuole mit kriiftiger that of plastid genomes3•4• Although the Wandung' (large vacuole 35-kb genome is distinct from the 6-7-kb with stout surrounds) ( see linear mitochondrial genome3-6, it is not refs cited in ref. 9). Our pre known where in the parasite cells the plas liminary experiments with tid-like genome resides. Plasmodium falciparum, the To determine whether a plastid is pre causative agent of the most sent, we used high-resolution in situ lethal form of malaria, hybridization7 to localize transcripts of a identify an organelle (not plastid-like 16S ribosomal RNA gene shown) which appears sim from Toxoplasma gondii8, the causative ilar to the T. gondii plastid. agent of toxoplasmosis. Transcripts accu The number of surrounding mulate in a small, ovoid organelle located membranes in the P. anterior to the nucleus in the mid-region falciparum plastid, and its of the cell (a, b in the figure). -

Membrane Structure and Function
Chapter 7 Membrane Structure and Function Lecture Outline Overview: Life at the Edge • The plasma membrane separates the living cell from its surroundings. • This thin barrier, 8 nm thick, controls traffic into and out of the cell. • Like all biological membranes, the plasma membrane is selectively permeable, allowing some substances to cross more easily than others. • The formation of a membrane that encloses a solution different from the surrounding solution while still permitting the uptake of nutrients and the elimination of waste products was a key event in the evolution of life. • The ability of the cell to discriminate in its chemical exchanges with its environment is fundamental to life. • It is the plasma membrane and its component molecules that make this selectivity possible. Concept 7.1 Cellular membranes are fluid mosaics of lipids and proteins. • The main macromolecules in membranes are lipids and proteins, but carbohydrates are also important. • The most abundant lipids are phospholipids. • Phospholipids and most other membrane constituents are amphipathic molecules, which have both hydrophobic and hydrophilic regions. Membrane models have evolved to fit new data. • The arrangement of phospholipids and proteins in biological membranes is described by the fluid mosaic model. • In this model, the membrane is a fluid structure with a “mosaic” of various proteins embedded in or attached to a double layer (bilayer) of phospholipids. • Models of membranes were developed long before membranes were first seen with electron microscopes in the 1950s. • In 1915, membranes isolated from red blood cells were chemically analyzed and found to be composed of lipids and proteins. • In 1925, E.