Supplementary Table 7 A. Niger CBS 513.88 Gene Expression Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -

Heterologous Expression and Identification of the Genes Involved
Heterologous Expression and Identification of the Genes Involved in Anaerobic Degradation of 1,3-Dihydroxybenzene (Resorcinol) in Azoarcus anaerobiusᰔ Paula I. Darley,1† Jutta A. Hellstern,1†‡ Javier I. Medina-Bellver,2 Silvia Marque´s,2 Bernhard Schink,1 and Bodo Philipp1* Fachbereich Biologie, Universita¨t Konstanz, D-78457 Constance, Germany,1 and Estacio´n Experimental del Zaidı´n, C/. Profesor Albareda 1, E-18008 Granada, Spain2 Received 9 November 2006/Accepted 9 March 2007 Azoarcus anaerobius, a strictly anaerobic, gram-negative bacterium, utilizes resorcinol as a sole carbon and energy source with nitrate as an electron acceptor. Previously, we showed that resorcinol degradation by this bacterium is initiated by two oxidative steps, both catalyzed by membrane-associated enzymes that lead to the formation of hydroxyhydroquinone (HHQ; 1,2,4-benzenetriol) and 2-hydroxy-1,4-benzoquinone (HBQ). This study presents evidence for the further degradation of HBQ in cell extracts to form acetic and malic acids. To identify the A. anaerobius genes required for anaerobic resorcinol catabolism, a cosmid library with genomic DNA was constructed and transformed into the phylogenetically related species Thauera aromatica, which cannot grow with resorcinol. By heterologous complementation, a transconjugant was identified that gained the ability to metabolize resorcinol. Its cosmid, designated R؉, carries a 29.88-kb chromosomal DNA fragment containing 22 putative genes. In cell extracts of T. aromatica transconjugants, resorcinol was degraded to HHQ, HBQ, and acetate, suggesting that cosmid R؉ carried all of the genes necessary for resorcinol degradation. On the basis of the physiological characterization of T. aromatica transconjugants carrying transposon insertions -in different genes of cosmid R؉, eight open reading frames were found to be essential for resorcinol miner alization. -

Metaproteogenomic Insights Beyond Bacterial Response to Naphthalene
ORIGINAL ARTICLE ISME Journal – Original article Metaproteogenomic insights beyond bacterial response to 5 naphthalene exposure and bio-stimulation María-Eugenia Guazzaroni, Florian-Alexander Herbst, Iván Lores, Javier Tamames, Ana Isabel Peláez, Nieves López-Cortés, María Alcaide, Mercedes V. del Pozo, José María Vieites, Martin von Bergen, José Luis R. Gallego, Rafael Bargiela, Arantxa López-López, Dietmar H. Pieper, Ramón Rosselló-Móra, Jesús Sánchez, Jana Seifert and Manuel Ferrer 10 Supporting Online Material includes Text (Supporting Materials and Methods) Tables S1 to S9 Figures S1 to S7 1 SUPPORTING TEXT Supporting Materials and Methods Soil characterisation Soil pH was measured in a suspension of soil and water (1:2.5) with a glass electrode, and 5 electrical conductivity was measured in the same extract (diluted 1:5). Primary soil characteristics were determined using standard techniques, such as dichromate oxidation (organic matter content), the Kjeldahl method (nitrogen content), the Olsen method (phosphorus content) and a Bernard calcimeter (carbonate content). The Bouyoucos Densimetry method was used to establish textural data. Exchangeable cations (Ca, Mg, K and 10 Na) extracted with 1 M NH 4Cl and exchangeable aluminium extracted with 1 M KCl were determined using atomic absorption/emission spectrophotometry with an AA200 PerkinElmer analyser. The effective cation exchange capacity (ECEC) was calculated as the sum of the values of the last two measurements (sum of the exchangeable cations and the exchangeable Al). Analyses were performed immediately after sampling. 15 Hydrocarbon analysis Extraction (5 g of sample N and Nbs) was performed with dichloromethane:acetone (1:1) using a Soxtherm extraction apparatus (Gerhardt GmbH & Co. -

Genome-Wide Transcriptional Changes and Lipid Profile
G C A T T A C G G C A T genes Article Genome-Wide Transcriptional Changes and Lipid Profile Modifications Induced by Medicago truncatula N5 Overexpression at an Early Stage of the Symbiotic Interaction with Sinorhizobium meliloti Chiara Santi 1, Barbara Molesini 1, Flavia Guzzo 1, Youry Pii 2 ID , Nicola Vitulo 1 and Tiziana Pandolfini 1,* ID 1 Department of Biotechnology, University of Verona, 37134 Verona, Italy; [email protected] (C.S.); [email protected] (B.M.); fl[email protected] (F.G.); [email protected] (N.V.) 2 Faculty of Science and Technology, Free University of Bozen-Bolzano, 39100 Bolzano BZ, Italy; [email protected] * Correspondence: tiziana.pandolfi[email protected]; Tel.: +39-045-8027918 Received: 30 October 2017; Accepted: 11 December 2017; Published: 19 December 2017 Abstract: Plant lipid-transfer proteins (LTPs) are small basic secreted proteins, which are characterized by lipid-binding capacity and are putatively involved in lipid trafficking. LTPs play a role in several biological processes, including the root nodule symbiosis. In this regard, the Medicago truncatula nodulin 5 (MtN5) LTP has been proved to positively regulate the nodulation capacity, controlling rhizobial infection and nodule primordia invasion. To better define the lipid transfer protein MtN5 function during the symbiosis, we produced MtN5-downregulated and -overexpressing plants, and we analysed the transcriptomic changes occurring in the roots at an early stage of Sinorhizobium meliloti infection. We also carried out the lipid profile analysis of wild type (WT) and MtN5-overexpressing roots after rhizobia infection. The downregulation of MtN5 increased the root hair curling, an early event of rhizobia infection, and concomitantly induced changes in the expression of defence-related genes. -

Rehmannia Glutinosa-Monocultured Rhizosphere Soil
Comparative Metaproteomic Analysis on Consecutively Rehmannia glutinosa-Monocultured Rhizosphere Soil Linkun Wu1,2, Haibin Wang1,2., Zhixing Zhang1,2., Rui Lin2,3, Zhongyi Zhang1,4, Wenxiong Lin1,2* 1 School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 2 Agroecological Institute, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 3 College of Oceanography and Environmental Science, Xiamen University, Xiamen, Fujian, China, 4 Institute of Chinese Medicinal Materials, Henan Agriculture University, Zhengzhou, Henan, China Abstract Background: The consecutive monoculture for most of medicinal plants, such as Rehmannia glutinosa, results in a significant reduction in the yield and quality. There is an urgent need to study for the sustainable development of Chinese herbaceous medicine. Methodology/Principal Findings: Comparative metaproteomics of rhizosphere soil was developed and used to analyze the underlying mechanism of the consecutive monoculture problems of R. glutinosa. The 2D-gel patterns of protein spots for the soil samples showed a strong matrix dependency. Among the spots, 103 spots with high resolution and repeatability were randomly selected and successfully identified by MALDI TOF-TOF MS for a rhizosphere soil metaproteomic profile analysis. These proteins originating from plants and microorganisms play important roles in nutrient cycles and energy flow in rhizospheric soil ecosystem. They function in protein, nucleotide and secondary metabolisms, signal transduction and resistance. Comparative metaproteomics analysis revealed 33 differentially expressed protein spots in rhizosphere soil in response to increasing years of monoculture. Among them, plant proteins related to carbon and nitrogen metabolism and stress response, were mostly up-regulated except a down-regulated protein (glutathione S-transferase) involving detoxification. -
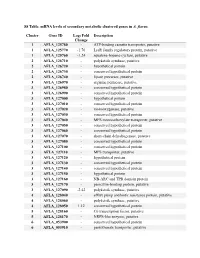
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

Table S1-Final.Xlsx
Table S1. Functional gene families covered on the GeoChip 5.0. No. of sequence‐ No. of group‐ No. of total No. of covered No. of total Category Subcategory Subcategory 2 Gene Encoded Enzyme specific probesa specific probesa probes on 5M CDSa probes on 5S Categories for Geochemical Cycling Carbon Cycling Carbon Degradation Agar beta_agarase Agarase 42 79 217 121 Agar Total 42 79 0 217 121 Alginate alginase Alginase 45 200 512 245 Alginate Total 45 200 0 512 245 Cellulose Endoglucanase Endoglucanase 198 248 446 740 343 Cellulose Total 198 248 446 740 343 Chitin Acetylglucosaminidase Acetylglucosaminidase 119 976 1095 3088 1116 Chitinase Chitinase 649 1231 1880 3285 1411 endochitinase Endochitinase 213 334 1104 547 exochitinase Exochitinase 14 55 259 69 Chitin Total 995 2596 2975 7736 3143 Glyoxylate cycle AceA Isocitrate lyase 69 373 442 887 0 AceB Malate synthase A 90 610 700 1457 0 Glyoxylate cycle Total 159 983 1142 2344 0 Hemicellulose Ara Arabinofuranosidase 172 543 715 1367 827 Mannanase Mannanase 159 238 397 639 478 Xylanase Xylanase 150 653 803 1739 858 Hemicellulose Total 481 1434 1915 3745 2163 Heparin heparinase Heparinase 7 53 166 60 Heparin Total 7 53 0 166 60 Hyaluronic acid hyaluronidase Hyaluronidase 880 383 88 Hyaluronidase Total 8 80 0 383 88 Lignin Glx Glyoxal oxidase 103 40 143 178 125 Mnp Manganese peroxidase 50 13 63 74 66 Phenol_oxidase Laccase or phenol oxidase 188 372 560 1009 677 Lignin Total 341 425 766 1261 868 Pectin Pectinase (pectate_lyase) Pectate lyase 53 252 305 827 321 Pme Pectin methylesterase 25 240 265 -

Association Between the Gut Microbiota and Blood Pressure in a Population Cohort of 6953 Individuals
Journal of the American Heart Association ORIGINAL RESEARCH Association Between the Gut Microbiota and Blood Pressure in a Population Cohort of 6953 Individuals Joonatan Palmu , MD; Aaro Salosensaari , MSc; Aki S. Havulinna , DSc (Tech); Susan Cheng , MD, MPH; Michael Inouye, PhD; Mohit Jain, MD, PhD; Rodolfo A. Salido , BSc; Karenina Sanders , BSc; Caitriona Brennan, BSc; Gregory C. Humphrey, BSc; Jon G. Sanders , PhD; Erkki Vartiainen , MD, PhD; Tiina Laatikainen , MD, PhD; Pekka Jousilahti, MD, PhD; Veikko Salomaa , MD, PhD; Rob Knight , PhD; Leo Lahti , DSc (Tech); Teemu J. Niiranen , MD, PhD BACKGROUND: Several small-scale animal studies have suggested that gut microbiota and blood pressure (BP) are linked. However, results from human studies remain scarce and conflicting. We wanted to elucidate the multivariable-adjusted as- sociation between gut metagenome and BP in a large, representative, well-phenotyped population sample. We performed a focused analysis to examine the previously reported inverse associations between sodium intake and Lactobacillus abun- dance and between Lactobacillus abundance and BP. METHODS AND RESULTS: We studied a population sample of 6953 Finns aged 25 to 74 years (mean age, 49.2±12.9 years; 54.9% women). The participants underwent a health examination, which included BP measurement, stool collection, and 24-hour urine sampling (N=829). Gut microbiota was analyzed using shallow shotgun metagenome sequencing. In age- and sex-adjusted models, the α (within-sample) and β (between-sample) diversities of taxonomic composition were strongly re- lated to BP indexes (P<0.001 for most). In multivariable-adjusted models, β diversity was only associated with diastolic BP (P=0.032). -

Phosphodiesterase 1B Knock-Out Mice Exhibit Exaggerated Locomotor Hyperactivity and DARPP-32 Phosphorylation in Response to Dopa
The Journal of Neuroscience, June 15, 2002, 22(12):5188–5197 Phosphodiesterase 1B Knock-Out Mice Exhibit Exaggerated Locomotor Hyperactivity and DARPP-32 Phosphorylation in Response to Dopamine Agonists and Display Impaired Spatial Learning Tracy M. Reed,1,3 David R. Repaske,2* Gretchen L. Snyder,4 Paul Greengard,4 and Charles V. Vorhees1* Divisions of 1Developmental Biology and 2Endocrinology, Children’s Hospital Research Foundation, Cincinnati, Ohio 45229, 3Department of Biology, College of Mount St. Joseph, Cincinnati, Ohio 45233, and 4Laboratory of Molecular and Cellular Neuroscience, Rockefeller University, New York, New York 10021 Using homologous recombination, we generated mice lack- maze spatial-learning deficits. These results indicate that en- ing phosphodiesterase-mediated (PDE1B) cyclic nucleotide- hancement of cyclic nucleotide signaling by inactivation of hydrolyzing activity. PDE1B Ϫ/Ϫ mice showed exaggerated PDE1B-mediated cyclic nucleotide hydrolysis plays a signifi- hyperactivity after acute D-methamphetamine administra- cant role in dopaminergic function through the DARPP-32 and tion. Striatal slices from PDE1B Ϫ/Ϫ mice exhibited increased related transduction pathways. levels of phospho-Thr 34 DARPP-32 and phospho-Ser 845 Key words: phosphodiesterases; DARPP-32; dopamine- GluR1 after dopamine D1 receptor agonist or forskolin stimu- stimulated locomotor activity; spatial learning and memory; lation. PDE1B Ϫ/Ϫ and PDE1B ϩ/Ϫ mice demonstrated Morris Morris water maze; methamphetamine; SKF81297; forskolin Calcium/calmodulin-dependent phosphodiesterases (CaM- (CaMKII) and calcineurin and have the potential to activate PDEs) are members of one of 11 families of PDEs (Soderling et CaM-PDEs. Dopamine D1 or D2 receptor activation leads to al., 1999;Yuasa et al., 2001) and comprise the only family that acts adenylyl cyclase activation or inhibition, respectively (Traficante ϩ as a potential point of interaction between the Ca 2 and cyclic et al., 1976; Monsma et al., 1990; Cunningham and Kelley, 1993; nucleotide signaling pathways. -

N-Glycan Trimming in the ER and Calnexin/Calreticulin Cycle
Neurotransmitter receptorsGABA and A postsynapticreceptor activation signal transmission Ligand-gated ion channel transport GABAGABA Areceptor receptor alpha-5 alpha-1/beta-1/gamma-2 subunit GABA A receptor alpha-2/beta-2/gamma-2GABA receptor alpha-4 subunit GABAGABA receptor A receptor beta-3 subunitalpha-6/beta-2/gamma-2 GABA-AGABA receptor; A receptor alpha-1/beta-2/gamma-2GABA receptoralpha-3/beta-2/gamma-2 alpha-3 subunit GABA-A GABAreceptor; receptor benzodiazepine alpha-6 subunit site GABA-AGABA-A receptor; receptor; GABA-A anion site channel (alpha1/beta2 interface) GABA-A receptor;GABA alpha-6/beta-3/gamma-2 receptor beta-2 subunit GABAGABA receptorGABA-A receptor alpha-2receptor; alpha-1 subunit agonist subunit GABA site Serotonin 3a (5-HT3a) receptor GABA receptorGABA-C rho-1 subunitreceptor GlycineSerotonin receptor subunit3 (5-HT3) alpha-1 receptor GABA receptor rho-2 subunit GlycineGlycine receptor receptor subunit subunit alpha-2 alpha-3 Ca2+ activated K+ channels Metabolism of ingested SeMet, Sec, MeSec into H2Se SmallIntermediateSmall conductance conductance conductance calcium-activated calcium-activated calcium-activated potassium potassium potassiumchannel channel protein channel protein 2 protein 1 4 Small conductance calcium-activatedCalcium-activated potassium potassium channel alpha/beta channel 1 protein 3 Calcium-activated potassiumHistamine channel subunit alpha-1 N-methyltransferase Neuraminidase Pyrimidine biosynthesis Nicotinamide N-methyltransferase Adenosylhomocysteinase PolymerasePolymeraseHistidine basic -

Reduction of Pectinesterase Activity in a Commercial Enzyme Preparation
Journal of the Science of Food and Agriculture J Sci Food Agric 85:1613–1621 (2005) DOI: 10.1002/jsfa.2154 Reduction of pectinesterase activity in a commercial enzyme preparation by pulsed electric fields: comparison of inactivation kinetic models Joaquın´ Giner, Pascal Grouberman, Vicente Gimeno and Olga Martın´ ∗ Department of Food Technology, University of Lleida, CeRTA-UTPV, ETSEA, Avda Alcalde Rovira Roure 191, 25198-Lleida, Spain Abstract: The inactivation of pectinesterase (PE) in a commercial enzyme preparation (CEP) under high intensity pulsed electric fields (HIPEF) was studied. After desalting and water dilution of the raw CEP, samples were exposed to exponentially decay waveform pulses for up to 463 µs at electric field intensities ranging from 19 to 38 kV cm−1. Pulses were applied in monopolar mode. Experimental data were fitted to a first-order kinetic model as well as to models based on Fermi, Hulsheger¨ or Weibull equations to describe PE inactivation kinetics. Characteristic parameters for each model were calculated. Relationships between some of the parameters and process variables were obtained. The Weibull model yielded the best accuracy factor. The relationship between residual PE and input of electrical energy density was found to be that of exponential decay. 2005 Society of Chemical Industry Keywords: pulsed electric fields; kinetics; pectinesterase; model; inactivation INTRODUCTION It has become customary to use CEPs in fruit and Pectinesterase (PE; EC 3.1.1.11) is a pectic enzyme vegetable juice technology. Depending -

Regulation of Calmodulin-Stimulated Cyclic Nucleotide Phosphodiesterase (PDE1): Review
95-105 5/6/06 13:44 Page 95 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 18: 95-105, 2006 95 Regulation of calmodulin-stimulated cyclic nucleotide phosphodiesterase (PDE1): Review RAJENDRA K. SHARMA, SHANKAR B. DAS, ASHAKUMARY LAKSHMIKUTTYAMMA, PONNIAH SELVAKUMAR and ANURAAG SHRIVASTAV Department of Pathology and Laboratory Medicine, College of Medicine, University of Saskatchewan, Cancer Research Division, Saskatchewan Cancer Agency, 20 Campus Drive, Saskatoon SK S7N 4H4, Canada Received January 16, 2006; Accepted March 13, 2006 Abstract. The response of living cells to change in cell 6. Differential inhibition of PDE1 isozymes and its environment depends on the action of second messenger therapeutic applications molecules. The two second messenger molecules cAMP and 7. Role of proteolysis in regulating PDE1A2 Ca2+ regulate a large number of eukaryotic cellular events. 8. Role of PDE1A1 in ischemic-reperfused heart Calmodulin-stimulated cyclic nucleotide phosphodiesterase 9. Conclusion (PDE1) is one of the key enzymes involved in the complex interaction between cAMP and Ca2+ second messenger systems. Some PDE1 isozymes have similar kinetic and 1. Introduction immunological properties but are differentially regulated by Ca2+ and calmodulin. Accumulating evidence suggests that the A variety of cellular activities are regulated through mech- activity of PDE1 is selectively regulated by cross-talk between anisms controlling the level of cyclic nucleotides. These Ca2+ and cAMP signalling pathways. These isozymes are mechanisms include synthesis, degradation, efflux and seque- also further distinguished by various pharmacological agents. stration of cyclic adenosine 3':5'-monophosphate (cAMP) and We have demonstrated a potentially novel regulation of PDE1 cyclic guanosine 3':5'- monophosphate (cGMP) within the by calpain.