The Rdna Loci—Intersections of Replication, Transcription, and Repair Pathways
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of a Non-LTR Retrotransposon from the Gypsy Moth
Insect Molecular Biology (1999) 8(2), 231-242 Identification of a non-L TR retrotransposon from the gypsy moth K. J. Garner and J. M. Siavicek sposons (Boeke & Corces, 1989), or retroposons USDA Forest Service, Northeastern Research Station, (McClure, 1991). Many non-L TR retrotransposons Delaware, Ohio, U.S.A. have been described in insects, including the Doc (O'Hare et al., 1991), F (Di Nocera & Casari, 1987), I (Fawcett et al., 1986) and jockey (Priimiigi et al., 1988) Abstract elements of Drosophila melanogaster, the T1Ag A family of highly repetitive elements, named LDT1, (Besansky, 1990) and Q (Besansky et al., 1994) ele- has been identified in the gypsy moth, Lymantria ments of Anopheles gambiae, and the R1Bm (Xiong & dispar. The complete element is 5.4 kb in length and Eickbush, 1988a) and R2Bm (Burke et al., 1987) lacks long-terminal repeats, The element contains two families of ribosomal DNA insertions in Bombyx mori. open reading frames with a significant amino acid Gypsy moths (Lymantria dispar) are currently wide- sequence similarity to several non-L TR retrotrans- spread forest pests in the north-eastern United States posons. The first open reading frame contains a and the adjacent regions of Canada. Population region that potentially encodes a polypeptide similar markers have been sought to distinguish the North to DNA-binding GAG-like proteins. The second American gypsy moths introduced from Europe in 1869 encodes a polypeptide resembling both endonuclease from those recently introduced from Asia (Bogdano- and reverse transcriptase sequences. A" members of wicz et al., 1993; Pfeifer et al., 1995; Garner & Siavicek, the LDT1 element family sequenced thus far have poly- 1996; Schreiber et al., 1997). -

1 Low Ribosomal RNA Genes Copy Number Provoke Genomic Instability
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.24.917823; this version posted January 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Low ribosomal RNA genes copy number provoke genomic instability and chromosomal segment duplication events that modify global gene expression and plant-pathogen response Ariadna Picart-Picolo1,2, Stefan Grob3, Nathalie Picault1,2, Michal Franek4, Thierry halter5, Tom R. Maier6, Christel Llauro1,2, Edouard Jobet1,2, Panpan Zhang1,2,7, Paramasivan Vijayapalani6, Thomas J. Baum6, Lionel Navarro5, Martina Dvorackova4, Marie Mirouze1,2,7, Frederic Pontvianne1,2# 1CNRS, LGDP UMR5096, Université de Perpignan, Perpignan, France 2UPVD, LGDP UMR5096, Université de Perpignan, Perpignan, France 3Institute of Plant and Microbial Biology, University of Zurich, Zurich, Switzerland 4 Mendel Centre for Plant Genomics and Proteomics, CEITEC, Masaryk University, Brno, Czech Republic 5ENS, IBENS, CNRS/INSERM, PSL Research University, Paris, France 6Department of Plant Pathology and Microbiology, Iowa State University, Ames, IA, USA 7IRD, UMR232 DIADE, Montpellier, France #corresponding author: [email protected] ABSTRACT Among the hundreds of ribosomal RNA (rRNA) gene copies organized as tandem repeats in the nucleolus organizer regions (NORs), only a portion is usually actively expressed in the nucleolus and participate in the ribosome biogenesis process. The role of these extra-copies remains elusive, but previous studies suggested their importance in genome stability and global 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.01.24.917823; this version posted January 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

The Activity and Evolution of the Daphnia Dna Transposon
THE ACTIVITY AND EVOLUTION OF THE DAPHNIA DNA TRANSPOSON POKEY A Thesis Presented to The Faculty of Graduate Studies of The University of Guelph by TYLER ADAM ELLIOTT In partial fulfilment of requirements for the degree of Master of Science January, 2011 © Tyler Adam Elliott, 2011 Library and Archives Bibliotheque et 1*1 Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington Ottawa ON K1A 0N4 Ottawa ON K1A 0N4 Canada Canada Your We Votre reference ISBN: 978-0-494-80087-4 Our file Notre r$f6rence ISBN: 978-0-494-80087-4 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library and permettant a la Bibliotheque et Archives Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par I'lnternet, preter, telecommunication or on the Internet, distribuer et vendre des theses partout dans le loan, distribute and sell theses monde, a des fins commerciales ou autres, sur worldwide, for commercial or non support microforme, papier, electronique et/ou commercial purposes, in microform, autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in this et des droits moraux qui protege cette these. Ni thesis. Neither the thesis nor la these ni des extraits substantiels de celle-ci substantial extracts from it may be ne doivent etre imprimes ou autrement printed or otherwise reproduced reproduits sans son autorisation. -

Ribosomal DNA Copy Loss and Repeat Instability in ATRX-Mutated Cancers
Ribosomal DNA copy loss and repeat instability in ATRX-mutated cancers Maheshi Udugamaa,1, Elaine Sanijb,c,1, Hsiao P. J. Voona, Jinbae Sonb, Linda Hiia, Jeremy D. Hensond, F. Lyn Chana, Fiona T. M. Changa, Yumei Liue, Richard B. Pearsona,b,f,g, Paul Kalitsish, Jeffrey R. Manni, Philippe Collasj,k, Ross D. Hannana,b,f,g,l, and Lee H. Wonga,2 aDepartment of Biochemistry and Molecular Biology, Monash Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia; bResearch Division, Peter MacCallum Cancer Centre, Parkville, VIC 2010, Australia; cDepartment of Pathology, The University of Melbourne, Parkville, VIC 3010, Australia; dCancer Cell Immortality Group, Adult Cancer Program, Prince of Wales Clinical School, University of New South Wales, Randwick, NSW 2052, Australia; eCollege of Animal Science and Technology, Henan University of Science and Technology, Luoyang, Henan Province, 471023, China; fSir Peter MacCallum Department of Oncology, The University of Melbourne, VIC 3010, Australia; gDepartment of Biochemistry and Molecular Biology, The University of Melbourne, Parkville, VIC 3010, Australia; hDepartment of Paediatrics, Murdoch Children’s Research Institute, Royal Children’s Hospital, University of Melbourne, Parkville, VIC 3052, Australia; iGenome Modification Platform, Monash University, Clayton, VIC 3800, Australia; jDepartment of Molecular Medicine, Institute of Basic Medical Sciences, Faculty of Medicine, University of Oslo, 0317 Oslo, Norway; kNorwegian Center for Stem Cell Research, Department of Immunology -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Retrotransposon R1bm Endonuclease Cleaves the Target Sequence
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 2083–2088, March 1998 Biochemistry Retrotransposon R1Bm endonuclease cleaves the target sequence QINGHUA FENG*†,GERALD SCHUMANN*, AND JEF D. BOEKE‡ Department of Molecular Biology and Genetics, The Johns Hopkins University School of Medicine, 725 North Wolfe Street, Baltimore MD 21205 Communicated by Thomas J. Kelly, The Johns Hopkins University, Baltimore, MD, December 23, 1997 (received for review July 4, 1997) ABSTRACT The R1Bm element, found in the silkworm Bombyx mori, is a member of a group of widely distributed retrotransposons that lack long terminal repeats. Some of these elements are highly sequence-specific and others, like the human L1 sequence, are less so. The majority of R1Bm elements are associated with ribosomal DNA (rDNA). R1Bm inserts into 28S rDNA at a specific sequence; after insertion it is flanked by a specific 14-bp target site duplication of the 28S rDNA. The basis for this sequence specificity is unknown. We show that R1Bm encodes an enzyme related to the endonuclease found in the human L1 retrotransposon and also to the apurinicyapyrimidinic endonucleases. We ex- pressed and purified the enzyme from bacteria and showed that it cleaves in vitro precisely at the positions in rDNA corresponding to the boundaries of the 14-bp target site duplication. We conclude that the function of the retrotrans- poson endonucleases is to define and cleave target site DNA. Retrotransposons that lack long terminal repeats are very diverse in structure and can insert into a wide variety of different types of DNA targets. Some of these elements, such as the human L1 element, insert into a relatively wide array of FIG. -

Regulation of Ribosomal DNA Amplification by the TOR Pathway
Regulation of ribosomal DNA amplification by the TOR pathway Carmen V. Jacka,1, Cristina Cruza,1, Ryan M. Hulla, Markus A. Kellerb, Markus Ralserb,c, and Jonathan Houseleya,2 aEpigenetics Programme, The Babraham Institute, Cambridge CB22 3AT, United Kingdom; bCambridge Systems Biology Centre and Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, United Kingdom; and cDivision of Physiology and Metabolism, Medical Research Council National Institute for Medical Research, London NW7 1AA, United Kingdom Edited by Jasper Rine, University of California, Berkeley, CA, and approved June 26, 2015 (received for review March 27, 2015) Repeated regions are widespread in eukaryotic genomes, and key The rate of recombination between copies is regulated by the functional elements such as the ribosomal DNA tend to be formed histone deacetylase (HDAC) Sir2 (11-13), and recombination of high copy repeated sequences organized in tandem arrays. In through this pathway is strictly dependent on the homologous general, high copy repeats are remarkably stable, but a number of recombination (HR) machinery (14). Frequent recombination organisms display rapid ribosomal DNA amplification at specific events are required to maintain rDNA homogeneity (15, 16) and times or under specific conditions. Here we demonstrate that result in the loss of markers integrated in the rDNA (7). However, target of rapamycin (TOR) signaling stimulates ribosomal DNA this HR-dependent pathway regulated by Sir2 is nondirectional; amplification in budding yeast, linking external nutrient avail- repeat gain and loss occurs at equivalent rates, so no change in ability to ribosomal DNA copy number. We show that ribosomal average copy number is observed over time (13). -

Organization and Evolution of 5S Ribosomal Dna in the Fish Genome
In: Focus on Genome Research ISSN 1-59033-960-6 Editor: Clyde R. Williams, pp335-363 ©2004 Nova Science Publishers, Inc. Chapter X ORGANIZATION AND EVOLUTION OF 5S RIBOSOMAL DNA IN THE FISH GENOME Cesar Martins 1 and Adriane Pinto Wasko 2 Departamento de Morfologia, Instituto de Biociências, Universidade Estadual Paulista, CEP 18618-000, Botucatu, SP, Brazil. Phone/Fax +55 14 38116264, 1e-mail [email protected]; 2e-mail: [email protected] ABSTRACT In higher eukaryotes, the 5S ribosomal multigene family (5S rDNA) is tandemly organized in repeat units composed of a coding region (5S rRNA gene) and a non-transcribed spacer sequence (NTS). Although the 5S rDNA organization has been investigated in several vertebrate species, present data are concentrated in mammals and amphibians, whereas other groups, such as fishes, have been poorly studied. To further the understanding on the dynamics and evolution of 5S rDNA arrays in the vertebrate genome, recent studies have focused on the genome organization of these sequences in fish species, which represent the base group of vertebrate evolution. It was evidenced that the chromosome distribution of the 5S rDNA is quite conserved among related fish species occupying an interstitial position in the chromosomes. Although the 5S rDNA clusters have been maintained conserved in the chromosomes, changes in the nucleotide sequences and organization of the repeat units have occurred in fish species, as demonstrated by the presence of 5S rDNA variant types within and between genomes clustered in distinct chromosome environments. These variants are distributed in two major classes, suggesting that such pattern could represent a primitive condition for the fish genome, as well as for vertebrates. -
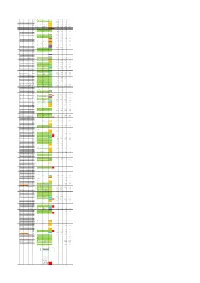
End Strand of Gene Gene Name Gene Function Starnd of Trascript
Genomic TSS strand Gene starnd of Transcipt relatively to TSS Start End Gene name coordinates relatively TSS type Control_fwd TEX_fwd Control_rev TEX_rev of gene function trascript gene group of TSS to ATG ribosomal 93 854 + rps12 + 1 protein S12 1 1 14.0638317 20.7539794 0 0 93 324 exon 1 + 111 rps12 intron 1 1 13.90756687 18.07149224 0.781323982 0.423550599 829 854 exon 2 + 496 rps12 exon2 2 24.22104343 15.24782157 0 0 30S 904 1371 + gene rps7 ribosomal + 2 protein S7 1303 -209 rps7 inter (ndhB) 2 20.47068832 9.035746118 0.625059185 0.847101199 NADH dehydrogen 1512 3535 + gene ndhB (ndh2) + ase subunit 3 2 1696 ndhB exon 1-inter 2 3.594090315 2.964854195 0.468794389 0.282367066 + 2209 ndhB exon 1-inter 2 46.09811492 4.09432246 0.468794389 0.423550599 1512 2237 exon 1 + 2756 ndhB exon 2 -inter 2 43.28534858 4.800240125 0.312529593 0.282367066 2756 3535 exon 2 + 3090 ndhB exon 2 -inter 2 17.50165719 15.95373924 0.312529593 0 + 3192 ndhB exon 2 -inter 2 140.6383167 117.6058831 2.812766334 1.694202397 - 3462 ndhB exon 2 -inter 3 1.406383167 1.129468265 1.406383167 3.67077186 4 3633 3712 + tRNA tRNA-Leu-CAA + 3610 -23 tRNA-Leu 1 77.19480938 84.85130339 0.625059185 0 + 3633 1 tRNA-Leu 1 359.5652963 649.0207016 0.781323982 0 photosyste 3954 4058 + gene psbM m II protein + 5 M 3775 -179 psbM 2 20.47068832 12.00060031 0 0.141183533 + 3954 psbM-0 2 69.22530477 28.37789015 0.156264796 0 hypothetical 4182 5073 + gene ycf66 6 protein 4182 4287 exon 1 4772 5073 exon 2 7 5202 5113 - gene ycf (ORF29) - 5299 orf29 inter 1 0 0 3.125295926 3.67077186 -

Intragenomic Heterogeneity of Intergenic Ribosomal DNA Spacers in Cucurbita Moschata Is Determined by DNA Minisatellites with Va
DNA Research, 2019, 26(3), 273–286 doi: 10.1093/dnares/dsz008 Advance Access Publication Date: 25 April 2019 Full Paper Full Paper Intragenomic heterogeneity of intergenic ribosomal DNA spacers in Cucurbita moschata is determined by DNA minisatellites with variable potential to form non-canonical DNA conformations Roman Matyasek* , Alena Kuderova, Eva Kutı´lkova, Marek Kucera, and Ales Kovarı´k Institute of Biophysics of the Czech Academy of Sciences, CZ-612 65 Brno, Czech Republic *To whom correspondence should be addressed. Tel. þ420 54 1517 230. Fax þ420 54 1211 293. Email: [email protected] Edited by Prof. Kazuhiro Sato Received 16 August 2018; Editorial decision 23 March 2019; Accepted 3 April 2019 Abstract The intergenic spacer (IGS) of rDNA is frequently built of long blocks of tandem repeats. To estimate the intragenomic variability of such knotty regions, we employed PacBio sequencing of the Cucurbita moschata genome, in which thousands of rDNA copies are distributed across a number of loci. The rRNA coding regions are highly conserved, indicating intensive interlocus homogenization and/or high selection pressure. However, the IGS exhibits high intragenomic structural diversity. Two repeated blocks, R1 (300–1250 bp) and R2 (290–643 bp), account for most of the IGS variation. They exhibit minisatellite-like features built of multiple periodically spaced short GC-rich sequence motifs with the potential to adopt non-canonical DNA conformations, G-quadruplex-folded and left-handed Z-DNA. The mutual arrangement of these motifs can be used to classify IGS variants into five structural families. Subtle polymorphisms exist within each family due to a variable number of repeats, suggesting the coexistence of an enormous number of IGS variants. -

Distribution of the DNA Transposon Family, Pokey in the Daphnia Pulex Species Complex Shannon H
Eagle and Crease Mobile DNA (2016) 7:11 DOI 10.1186/s13100-016-0067-7 RESEARCH Open Access Distribution of the DNA transposon family, Pokey in the Daphnia pulex species complex Shannon H. C. Eagle and Teresa J. Crease* Abstract Background: The Pokey family of DNA transposons consists of two putatively autonomous groups, PokeyAandPokeyB, and two groups of Miniature Inverted-repeat Transposable Elements (MITEs), mPok1andmPok2. This TE family is unusual as it inserts into a specific site in ribosomal (r)DNA, as well as other locations in Daphnia genomes. The goals of this study were to determine the distribution of the Pokey family in lineages of the Daphnia pulex species complex, and to test the hypothesis that unusally high PokeyAnumberinsomeisolatesofDaphnia pulicaria is the result of recent transposition. To do this, we estimated the haploid number of Pokey, mPok, and rRNA genes in 45 isolates from five Daphnia lineages using quantitative PCR. We also cloned and sequenced partial copies of PokeyAfromfourisolatesofD. pulicaria. Results: Haploid PokeyAandPokeyB number is generally less than 20 and tends to be higher outside rDNA in four lineages. Conversely, the number of both groups is much higher outside rDNA (~120) in D. arenata,andPokeyBisalso somewhat higher inside rDNA. mPok1 was only detected in D. arenata. mPok2occursbothoutside(~30)andinsiderDNA (~6) in D. arenata,butwasrare(≤2) outside rDNA in the other four lineages. There is no correlation between Pokey and rRNA gene number (mean = 240 across lineages) in any lineage. Variation among cloned partial PokeyAsequencesis significantly higher in isolates with high number compared to isolates with an average number. Conclusions: The high Pokey number outside rDNA in D. -

Phenotypic and Genotypic Consequences of CRISPR/Cas9
| INVESTIGATION Phenotypic and Genotypic Consequences of CRISPR/ Cas9 Editing of the Replication Origins in the rDNA of Saccharomyces cerevisiae Joseph C. Sanchez,*,†,‡,1 Anja Ollodart,*,† Christopher R. L. Large,*,† Courtnee Clough,*,† Gina M. Alvino,* Mitsuhiro Tsuchiya,§ Matthew Crane,§ Elizabeth X. Kwan,* Matt Kaeberlein,*,†,§ Maitreya J. Dunham,*,† M. K. Raghuraman,* and Bonita J. Brewer*,†,1,2 *Department of Genome Sciences, †Molecular and Cellular Biology Program, and §Department of Pathology, University of Washington, Seattle, Washington 98195 and ‡Bioscience Division, Los Alamos National Laboratory, Los Alamos, New Mexico 87544 ORCID IDs: 0000-00002-2444-1834 (J.C.S.); 0000-0002-6329-9463 (A.O.); 0000-0002-5732-970X (C.R.L.); 0000-0002-7859-7252 (C.C.); 0000-0002- 1311-3421 (M.K.); 0000-0001-9944-2666 (M.J.D.); 0000-0003-2382-3707 (M.K.R.); 0000-0001-8782-0471 (B.J.B.) ABSTRACT The complex structure and repetitive nature of eukaryotic ribosomal DNA (rDNA) is a challenge for genome assembly, thus the consequences of sequence variation in rDNA remain unexplored. However, renewed interest in the role that rDNA variation may play in diverse cellular functions, aside from ribosome production, highlights the need for a method that would permit genetic manipulation of the rDNA. Here, we describe a clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-based strategy to edit the rDNA locus in the budding yeast Saccharomyces cerevisiae, developed independently but similar to one developed by others. Using this approach, we modified the endogenous rDNA origin of replication in each repeat by deleting or replacing its consensus sequence. We characterized the transformants that have successfully modified their rDNA locus and propose a mechanism for how CRISPR/Cas9-mediated editing of the rDNA occurs.