Ribosomal DNA Copy Loss and Repeat Instability in ATRX-Mutated Cancers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of a Non-LTR Retrotransposon from the Gypsy Moth
Insect Molecular Biology (1999) 8(2), 231-242 Identification of a non-L TR retrotransposon from the gypsy moth K. J. Garner and J. M. Siavicek sposons (Boeke & Corces, 1989), or retroposons USDA Forest Service, Northeastern Research Station, (McClure, 1991). Many non-L TR retrotransposons Delaware, Ohio, U.S.A. have been described in insects, including the Doc (O'Hare et al., 1991), F (Di Nocera & Casari, 1987), I (Fawcett et al., 1986) and jockey (Priimiigi et al., 1988) Abstract elements of Drosophila melanogaster, the T1Ag A family of highly repetitive elements, named LDT1, (Besansky, 1990) and Q (Besansky et al., 1994) ele- has been identified in the gypsy moth, Lymantria ments of Anopheles gambiae, and the R1Bm (Xiong & dispar. The complete element is 5.4 kb in length and Eickbush, 1988a) and R2Bm (Burke et al., 1987) lacks long-terminal repeats, The element contains two families of ribosomal DNA insertions in Bombyx mori. open reading frames with a significant amino acid Gypsy moths (Lymantria dispar) are currently wide- sequence similarity to several non-L TR retrotrans- spread forest pests in the north-eastern United States posons. The first open reading frame contains a and the adjacent regions of Canada. Population region that potentially encodes a polypeptide similar markers have been sought to distinguish the North to DNA-binding GAG-like proteins. The second American gypsy moths introduced from Europe in 1869 encodes a polypeptide resembling both endonuclease from those recently introduced from Asia (Bogdano- and reverse transcriptase sequences. A" members of wicz et al., 1993; Pfeifer et al., 1995; Garner & Siavicek, the LDT1 element family sequenced thus far have poly- 1996; Schreiber et al., 1997). -

What Are Their Roles in Mitochondrial Protein Synthesis?
Characterisation of human mtRF1 and C12orf65: What are their roles in mitochondrial protein synthesis? Aleksandra Pajak M.Res Thesis submitted to Newcastle University in candidature for the degree of Doctor of Philosophy Newcastle University Faculty of Medical Sciences Institute for Ageing and Health Mitochondrial Research Group January 2013 Abstract Mitochondria have their own protein synthesis machinery that synthesises the oxidative phosphorylation components encoded by their mtDNA. This translation process consists of four main phases: initiation, elongation, termination and ribosome recycling. Termination and its control have been the least investigated. Recently, however, the termination factor, mtRF1a, has been characterised as sufficient to release all the nascent proteins from the mitoribosome. Furthermore, bioinformatics has identified three additional members of this mitochondrial release factor family namely, mtRF1, C12orf65 and ICT1. The latter is now known to be incorporated into the mitoribosome but its exact function remains unclear. My project has therefore focussed on characterising the remaining two factors; mtRF1 and C12orf65, and investigating their possible involvement in mitochondrial protein synthesis. It has been demonstrated that protein synthesis is not perfect and bacterial ribosomes not infrequently stall during translation. This can result from limiting amounts of charged tRNAs, stable secondary structures, or truncated/degraded transcripts. Ribosome stalling has been shown to cause growth arrest. In order to prevent that and maintain high efficiency of mitochondrial protein synthesis such stalled complexes need to be rapidly recycled. Bacteria have developed at least three distinct mechanisms by which ribosomes can be rescued. Contrastingly, despite the presence of truncated mRNAs in mitochondria, no such quality control mechanisms have been identified in these organelles. -

Analysis of the Relationship Between Ribosomal Protein and SSU Processome Assembly in Saccharomyces Cerevisiae
Analysis of the relationship between ribosomal protein and SSU processome assembly in Saccharomyces cerevisiae Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften (Dr. rer. nat.) der naturwissenschaftlichen Fakultät III – Biologie und vorklinische Medizin - der Universität Regensburg vorgelegt von Steffen Jakob aus Wolfen Januar 2010 Promotionsgesuch eingereicht am: 13. Januar 2010 Die Arbeit wurde angeleitet von: Prof. Dr. Herbert Tschochner Prüfungsausschuss: Vorsitzender: Prof. Dr. Armin Kurtz 1. Prüfer: Prof. Dr. Herbert Tschochner 2. Prüfer: Prof. Dr. Rainer Deutzmann 3. Prüfer: Prof. Dr. Wolfgang Seufert Tag der mündlichen Prüfung: 24. März 2010 Die vorliegende Arbeit wurde in der Zeit von April 2006 bis Januar 2010 am Lehrstuhl Biochemie III des Institutes für Biochemie, Genetik und Mikrobiologie der Naturwissenschaftlichen Fakultät III der Universität zu Regensburg unter Anleitung von Dr. Philipp Milkereit im Labor von Prof. Dr. Herbert Tschochner angefertigt. Ich erkläre hiermit, dass ich diese Arbeit selbst verfasst und keine anderen als die angegebenen Quellen und Hilfsmittel verwendet habe. Diese Arbeit war bisher noch nicht Bestandteil eines Prüfungsverfahrens. Andere Promotionsversuche wurden nicht unternommen. Regensburg, den 13. Januar 2010 Steffen Jakob Table of Contents Table of Contents 1 SUMMARY ...................................................................................................... 1 2 INTRODUCTION ............................................................................................ -

1 Low Ribosomal RNA Genes Copy Number Provoke Genomic Instability
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.24.917823; this version posted January 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Low ribosomal RNA genes copy number provoke genomic instability and chromosomal segment duplication events that modify global gene expression and plant-pathogen response Ariadna Picart-Picolo1,2, Stefan Grob3, Nathalie Picault1,2, Michal Franek4, Thierry halter5, Tom R. Maier6, Christel Llauro1,2, Edouard Jobet1,2, Panpan Zhang1,2,7, Paramasivan Vijayapalani6, Thomas J. Baum6, Lionel Navarro5, Martina Dvorackova4, Marie Mirouze1,2,7, Frederic Pontvianne1,2# 1CNRS, LGDP UMR5096, Université de Perpignan, Perpignan, France 2UPVD, LGDP UMR5096, Université de Perpignan, Perpignan, France 3Institute of Plant and Microbial Biology, University of Zurich, Zurich, Switzerland 4 Mendel Centre for Plant Genomics and Proteomics, CEITEC, Masaryk University, Brno, Czech Republic 5ENS, IBENS, CNRS/INSERM, PSL Research University, Paris, France 6Department of Plant Pathology and Microbiology, Iowa State University, Ames, IA, USA 7IRD, UMR232 DIADE, Montpellier, France #corresponding author: [email protected] ABSTRACT Among the hundreds of ribosomal RNA (rRNA) gene copies organized as tandem repeats in the nucleolus organizer regions (NORs), only a portion is usually actively expressed in the nucleolus and participate in the ribosome biogenesis process. The role of these extra-copies remains elusive, but previous studies suggested their importance in genome stability and global 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.01.24.917823; this version posted January 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -
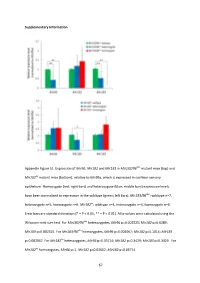
And Mir183 in Mir183/96 Dko Mutant Mice (Top) And
Supplementary Information Appendix Figure S1. Expression of Mir96 , Mir182 and Mir183 in Mir183/96 dko mutant mice (top) and Mir182 ko mutant mice (bottom), relative to Mir99a , which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96 dko : wildtype n=7, heterozygote n=5, homozygote n=6. Mir182 ko : wildtype n=4, heterozygote n=4, homozygote n=4. Error bars are standard deviation (* = P < 0.05, ** = P < 0.01). All p-values were calculated using the Wilcoxon rank sum test. For Mir183/96 dko heterozygotes, Mir96 p=0.002525; Mir182 p=0.6389; Mir183 p=0.002525. For Mir183/96 dko homozygotes, Mir96 p=0.002067; Mir182 p=0.1014; Mir183 p=0.002067. For Mir182 ko heterozygotes, Mir96 p=0.05714; Mir182 p=0.3429; Mir183 p=0.3429. For Mir182 ko homozygotes, Mir96 p=1; Mir182 p=0.02652; Mir183 p=0.05714. 67 68 Appendix Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96 dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 69 70 Appendix Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182 ko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 71 Appendix Figure S4. Mean ABR waveforms at 12kHz, shown at 20dB (top) and 50dB (bottom) above threshold (sensation level, SL) ± standard deviation, at four weeks old. -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

The Activity and Evolution of the Daphnia Dna Transposon
THE ACTIVITY AND EVOLUTION OF THE DAPHNIA DNA TRANSPOSON POKEY A Thesis Presented to The Faculty of Graduate Studies of The University of Guelph by TYLER ADAM ELLIOTT In partial fulfilment of requirements for the degree of Master of Science January, 2011 © Tyler Adam Elliott, 2011 Library and Archives Bibliotheque et 1*1 Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington Ottawa ON K1A 0N4 Ottawa ON K1A 0N4 Canada Canada Your We Votre reference ISBN: 978-0-494-80087-4 Our file Notre r$f6rence ISBN: 978-0-494-80087-4 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library and permettant a la Bibliotheque et Archives Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par I'lnternet, preter, telecommunication or on the Internet, distribuer et vendre des theses partout dans le loan, distribute and sell theses monde, a des fins commerciales ou autres, sur worldwide, for commercial or non support microforme, papier, electronique et/ou commercial purposes, in microform, autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in this et des droits moraux qui protege cette these. Ni thesis. Neither the thesis nor la these ni des extraits substantiels de celle-ci substantial extracts from it may be ne doivent etre imprimes ou autrement printed or otherwise reproduced reproduits sans son autorisation. -

Micrornas Mediated Regulation of the Ribosomal Proteins and Its Consequences on the Global Translation of Proteins
cells Review microRNAs Mediated Regulation of the Ribosomal Proteins and Its Consequences on the Global Translation of Proteins Abu Musa Md Talimur Reza 1,2 and Yu-Guo Yuan 1,3,* 1 Jiangsu Co-Innovation Center of Prevention and Control of Important Animal Infectious Diseases and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou 225009, China; [email protected] 2 Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawi´nskiego5a, 02-106 Warsaw, Poland 3 Jiangsu Key Laboratory of Zoonosis/Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou University, Yangzhou 225009, China * Correspondence: [email protected]; Tel.: +86-514-8797-9228 Abstract: Ribosomal proteins (RPs) are mostly derived from the energy-consuming enzyme families such as ATP-dependent RNA helicases, AAA-ATPases, GTPases and kinases, and are important structural components of the ribosome, which is a supramolecular ribonucleoprotein complex, composed of Ribosomal RNA (rRNA) and RPs, coordinates the translation and synthesis of proteins with the help of transfer RNA (tRNA) and other factors. Not all RPs are indispensable; in other words, the ribosome could be functional and could continue the translation of proteins instead of lacking in some of the RPs. However, the lack of many RPs could result in severe defects in the biogenesis of ribosomes, which could directly influence the overall translation processes and global expression of the proteins leading to the emergence of different diseases including cancer. While microRNAs (miRNAs) are small non-coding RNAs and one of the potent regulators of the post-transcriptional 0 gene expression, miRNAs regulate gene expression by targeting the 3 untranslated region and/or coding region of the messenger RNAs (mRNAs), and by interacting with the 50 untranslated region, Citation: Reza, A.M.M.T.; Yuan, Y.-G. -

Ribosomal DNA Copy Loss and Repeat Instability in ATRX-Mutated Cancers
Ribosomal DNA copy loss and repeat instability in ATRX-mutated cancers Maheshi Udugamaa,1, Elaine Sanijb,c,1, Hsiao P. J. Voona, Jinbae Sonb, Linda Hiia, Jeremy D. Hensond, F. Lyn Chana, Fiona T. M. Changa, Yumei Liue, Richard B. Pearsona,b,f,g, Paul Kalitsish, Jeffrey R. Manni, Philippe Collasj,k, Ross D. Hannana,b,f,g,l, and Lee H. Wonga,2 aDepartment of Biochemistry and Molecular Biology, Monash Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia; bResearch Division, Peter MacCallum Cancer Centre, Parkville, VIC 2010, Australia; cDepartment of Pathology, The University of Melbourne, Parkville, VIC 3010, Australia; dCancer Cell Immortality Group, Adult Cancer Program, Prince of Wales Clinical School, University of New South Wales, Randwick, NSW 2052, Australia; eCollege of Animal Science and Technology, Henan University of Science and Technology, Luoyang, Henan Province, 471023, China; fSir Peter MacCallum Department of Oncology, The University of Melbourne, VIC 3010, Australia; gDepartment of Biochemistry and Molecular Biology, The University of Melbourne, Parkville, VIC 3010, Australia; hDepartment of Paediatrics, Murdoch Children’s Research Institute, Royal Children’s Hospital, University of Melbourne, Parkville, VIC 3052, Australia; iGenome Modification Platform, Monash University, Clayton, VIC 3800, Australia; jDepartment of Molecular Medicine, Institute of Basic Medical Sciences, Faculty of Medicine, University of Oslo, 0317 Oslo, Norway; kNorwegian Center for Stem Cell Research, Department of Immunology -

History of the Ribosome and the Origin of Translation
History of the ribosome and the origin of translation Anton S. Petrova,1, Burak Gulena, Ashlyn M. Norrisa, Nicholas A. Kovacsa, Chad R. Berniera, Kathryn A. Laniera, George E. Foxb, Stephen C. Harveyc, Roger M. Wartellc, Nicholas V. Huda, and Loren Dean Williamsa,1 aSchool of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, GA 30332; bDepartment of Biology and Biochemistry, University of Houston, Houston, TX, 77204; and cSchool of Biology, Georgia Institute of Technology, Atlanta, GA 30332 Edited by David M. Hillis, The University of Texas at Austin, Austin, TX, and approved November 6, 2015 (received for review May 18, 2015) We present a molecular-level model for the origin and evolution of building up of the functional centers, proceeds to the establishment the translation system, using a 3D comparative method. In this model, of the common core, and continues to the development of large the ribosome evolved by accretion, recursively adding expansion metazoan rRNAs. segments, iteratively growing, subsuming, and freezing the rRNA. Incremental evolution of function is mapped out by stepwise Functions of expansion segments in the ancestral ribosome are accretion of rRNA. In the extant ribosome, specific segments of assigned by correspondence with their functions in the extant rRNA perform specific functions including peptidyl transfer, ribosome. The model explains the evolution of the large ribosomal subunit association, decoding, and energy-driven translocation subunit, the small ribosomal subunit, tRNA, and mRNA. Prokaryotic (11). The model assumes that the correlations of rRNA segments ribosomes evolved in six phases, sequentially acquiring capabilities with their functions have been reasonably maintained over the for RNA folding, catalysis, subunit association, correlated evolution, broad course of ribosomal evolution. -

Retrotransposon R1bm Endonuclease Cleaves the Target Sequence
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 2083–2088, March 1998 Biochemistry Retrotransposon R1Bm endonuclease cleaves the target sequence QINGHUA FENG*†,GERALD SCHUMANN*, AND JEF D. BOEKE‡ Department of Molecular Biology and Genetics, The Johns Hopkins University School of Medicine, 725 North Wolfe Street, Baltimore MD 21205 Communicated by Thomas J. Kelly, The Johns Hopkins University, Baltimore, MD, December 23, 1997 (received for review July 4, 1997) ABSTRACT The R1Bm element, found in the silkworm Bombyx mori, is a member of a group of widely distributed retrotransposons that lack long terminal repeats. Some of these elements are highly sequence-specific and others, like the human L1 sequence, are less so. The majority of R1Bm elements are associated with ribosomal DNA (rDNA). R1Bm inserts into 28S rDNA at a specific sequence; after insertion it is flanked by a specific 14-bp target site duplication of the 28S rDNA. The basis for this sequence specificity is unknown. We show that R1Bm encodes an enzyme related to the endonuclease found in the human L1 retrotransposon and also to the apurinicyapyrimidinic endonucleases. We ex- pressed and purified the enzyme from bacteria and showed that it cleaves in vitro precisely at the positions in rDNA corresponding to the boundaries of the 14-bp target site duplication. We conclude that the function of the retrotrans- poson endonucleases is to define and cleave target site DNA. Retrotransposons that lack long terminal repeats are very diverse in structure and can insert into a wide variety of different types of DNA targets. Some of these elements, such as the human L1 element, insert into a relatively wide array of FIG. -

Regulation of Ribosomal DNA Amplification by the TOR Pathway
Regulation of ribosomal DNA amplification by the TOR pathway Carmen V. Jacka,1, Cristina Cruza,1, Ryan M. Hulla, Markus A. Kellerb, Markus Ralserb,c, and Jonathan Houseleya,2 aEpigenetics Programme, The Babraham Institute, Cambridge CB22 3AT, United Kingdom; bCambridge Systems Biology Centre and Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, United Kingdom; and cDivision of Physiology and Metabolism, Medical Research Council National Institute for Medical Research, London NW7 1AA, United Kingdom Edited by Jasper Rine, University of California, Berkeley, CA, and approved June 26, 2015 (received for review March 27, 2015) Repeated regions are widespread in eukaryotic genomes, and key The rate of recombination between copies is regulated by the functional elements such as the ribosomal DNA tend to be formed histone deacetylase (HDAC) Sir2 (11-13), and recombination of high copy repeated sequences organized in tandem arrays. In through this pathway is strictly dependent on the homologous general, high copy repeats are remarkably stable, but a number of recombination (HR) machinery (14). Frequent recombination organisms display rapid ribosomal DNA amplification at specific events are required to maintain rDNA homogeneity (15, 16) and times or under specific conditions. Here we demonstrate that result in the loss of markers integrated in the rDNA (7). However, target of rapamycin (TOR) signaling stimulates ribosomal DNA this HR-dependent pathway regulated by Sir2 is nondirectional; amplification in budding yeast, linking external nutrient avail- repeat gain and loss occurs at equivalent rates, so no change in ability to ribosomal DNA copy number. We show that ribosomal average copy number is observed over time (13).