Produktinformation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NATURAL KILLER CELLS, HYPOXIA, and EPIGENETIC REGULATION of HEMOCHORIAL PLACENTATION by Damayanti Chakraborty Submitted to the G
NATURAL KILLER CELLS, HYPOXIA, AND EPIGENETIC REGULATION OF HEMOCHORIAL PLACENTATION BY Damayanti Chakraborty Submitted to the graduate degree program in Pathology and Laboratory Medicine and the Graduate Faculty of the University of Kansas in partial fulfillment ofthe requirements for the degree of Doctor of Philosophy. ________________________________ Chair: Michael J. Soares, Ph.D. ________________________________ Jay Vivian, Ph.D. ________________________________ Patrick Fields, Ph.D. ________________________________ Soumen Paul, Ph.D. ________________________________ Michael Wolfe, Ph.D. ________________________________ Adam J. Krieg, Ph.D. Date Defended: 04/01/2013 The Dissertation Committee for Damayanti Chakraborty certifies that this is the approved version of the following dissertation: NATURAL KILLER CELLS, HYPOXIA, AND EPIGENETIC REGULATION OF HEMOCHORIAL PLACENTATION ________________________________ Chair: Michael J. Soares, Ph.D. Date approved: 04/01/2013 ii ABSTRACT During the establishment of pregnancy, uterine stromal cells differentiate into decidual cells and recruit natural killer (NK) cells. These NK cells are characterized by low cytotoxicity and distinct cytokine production. In rodent as well as in human pregnancy, the uterine NK cells peak in number around mid-gestation after which they decline. NK cells associate with uterine spiral arteries and are implicated in pregnancy associated vascular remodeling processes and potentially in modulating trophoblast invasion. Failure of trophoblast invasion and vascular remodeling has been shown to be associated with pathological conditions like preeclampsia syndrome, hypertension in mother and/or fetal growth restriction. We hypothesize that NK cells fundamentally contribute to the organization of the placentation site. In order to study the in vivo role of NK cells during pregnancy, gestation stage- specific NK cell depletion was performed in rats using anti asialo GM1 antibodies. -

Antigen-Specific Memory CD4 T Cells Coordinated Changes in DNA
Downloaded from http://www.jimmunol.org/ by guest on September 24, 2021 is online at: average * The Journal of Immunology The Journal of Immunology published online 18 March 2013 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2013/03/17/jimmun ol.1202267 Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto, Katsumi Ogoshi, Atsushi Sasaki, Jun Abe, Wei Qu, Yoichiro Nakatani, Budrul Ahsan, Kenshiro Oshima, Francis H. W. Shand, Akio Ametani, Yutaka Suzuki, Shuichi Kaneko, Takashi Wada, Masahira Hattori, Sumio Sugano, Shinichi Morishita and Kouji Matsushima J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Author Choice option Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Freely available online through http://www.jimmunol.org/content/suppl/2013/03/18/jimmunol.120226 7.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Author Choice Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 24, 2021. Published March 18, 2013, doi:10.4049/jimmunol.1202267 The Journal of Immunology Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto,*,†,‡ Katsumi Ogoshi,* Atsushi Sasaki,† Jun Abe,* Wei Qu,† Yoichiro Nakatani,† Budrul Ahsan,x Kenshiro Oshima,† Francis H. -

The Genetic Program of Pancreatic Beta-Cell Replication in Vivo
Page 1 of 65 Diabetes The genetic program of pancreatic beta-cell replication in vivo Agnes Klochendler1, Inbal Caspi2, Noa Corem1, Maya Moran3, Oriel Friedlich1, Sharona Elgavish4, Yuval Nevo4, Aharon Helman1, Benjamin Glaser5, Amir Eden3, Shalev Itzkovitz2, Yuval Dor1,* 1Department of Developmental Biology and Cancer Research, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 2Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. 3Department of Cell and Developmental Biology, The Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem 91904, Israel 4Info-CORE, Bioinformatics Unit of the I-CORE Computation Center, The Hebrew University and Hadassah, The Institute for Medical Research Israel- Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 5Endocrinology and Metabolism Service, Department of Internal Medicine, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel *Correspondence: [email protected] Running title: The genetic program of pancreatic β-cell replication 1 Diabetes Publish Ahead of Print, published online March 18, 2016 Diabetes Page 2 of 65 Abstract The molecular program underlying infrequent replication of pancreatic beta- cells remains largely inaccessible. Using transgenic mice expressing GFP in cycling cells we sorted live, replicating beta-cells and determined their transcriptome. Replicating beta-cells upregulate hundreds of proliferation- related genes, along with many novel putative cell cycle components. Strikingly, genes involved in beta-cell functions, namely glucose sensing and insulin secretion were repressed. Further studies using single molecule RNA in situ hybridization revealed that in fact, replicating beta-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in beta-cell function. -

An Arabidopsis Nucleoporin NUP85 Modulates Plant Responses to ABA and Salt Stress
RESEARCH ARTICLE An Arabidopsis Nucleoporin NUP85 modulates plant responses to ABA and salt stress Yingfang Zhu1,2*, Bangshing Wang2, Kai Tang1,2, Chuan-Chih Hsu3, Shaojun Xie1,2, Hai Du2,4, Yuting Yang2,5, Weiguo Andy Tao3, Jian-Kang Zhu1,2* 1 Shanghai Center for Plant Stress Biology and CAS Center of Excellence for Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai, China, 2 Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN, United States of America, 3 Department of Biochemistry, Purdue University, West Lafayette, IN, United States of America, 4 College of Agronomy and Biotechnology, Southwest University, Chongqing, China, 5 Key Laboratory of Sugarcane Biology and Genetic Breeding Ministry of Agriculture, Fujian Agriculture and Forestry University Fuzhou, Fuzhou, China a1111111111 a1111111111 * [email protected] (YZ); [email protected] (JKZ) a1111111111 a1111111111 a1111111111 Abstract Several nucleoporins in the nuclear pore complex (NPC) have been reported to be involved in abiotic stress responses in plants. However, the molecular mechanism of how NPC regu- lates abiotic stress responses, especially the expression of stress responsive genes OPEN ACCESS remains poorly understood. From a forward genetics screen using an abiotic stress-respon- Citation: Zhu Y, Wang B, Tang K, Hsu C-C, Xie S, Du H, et al. (2017) An Arabidopsis Nucleoporin sive luciferase reporter (RD29A-LUC) in the sickle-1 (sic-1) mutant background, we identi- NUP85 modulates plant responses to ABA and salt fied a suppressor caused by a mutation in NUCLEOPORIN 85 (NUP85), which exhibited stress. PLoS Genet 13(12): e1007124. https://doi. reduced expression of RD29A-LUC in response to ABA and salt stress. -

Induction of Therapeutic Tissue Tolerance Foxp3 Expression Is
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2012; 189:3947-3956; Prepublished online 17 from submission to initial decision 4 weeks from acceptance to publication September 2012; doi: 10.4049/jimmunol.1200449 http://www.jimmunol.org/content/189/8/3947 Foxp3 Expression Is Required for the Induction of Therapeutic Tissue Tolerance Frederico S. Regateiro, Ye Chen, Adrian R. Kendal, Robert Hilbrands, Elizabeth Adams, Stephen P. Cobbold, Jianbo Ma, Kristian G. Andersen, Alexander G. Betz, Mindy Zhang, Shruti Madhiwalla, Bruce Roberts, Herman Waldmann, Kathleen F. Nolan and Duncan Howie J Immunol cites 35 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/09/17/jimmunol.120044 9.DC1 This article http://www.jimmunol.org/content/189/8/3947.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. -

Plasticity and the Impact of Increasing Temperature on a Tropical Ectotherm
Georgia Southern University Digital Commons@Georgia Southern Electronic Theses and Dissertations Graduate Studies, Jack N. Averitt College of Spring 2020 Plasticity and the Impact of Increasing Temperature on a Tropical Ectotherm Adam A. Rosso Follow this and additional works at: https://digitalcommons.georgiasouthern.edu/etd Part of the Bioinformatics Commons, Comparative and Evolutionary Physiology Commons, Evolution Commons, Integrative Biology Commons, Other Ecology and Evolutionary Biology Commons, and the Other Genetics and Genomics Commons Recommended Citation Rosso, Adam A., "Plasticity and the Impact of Increasing Temperature on a Tropical Ectotherm" (2020). Electronic Theses and Dissertations. 2069. https://digitalcommons.georgiasouthern.edu/etd/2069 This thesis (open access) is brought to you for free and open access by the Graduate Studies, Jack N. Averitt College of at Digital Commons@Georgia Southern. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Digital Commons@Georgia Southern. For more information, please contact [email protected]. PLASTICITY AND THE IMPAPCT OF INCREASING TEMPERATURE ON A TROPICAL ECTOTHERM by ADAM A. ROSSO (Under the direction of Christian L. Cox) ABSTRACT Organisms may respond to climate change through behavior, genetic adaptation, and/or phenotypic plasticity. Tropical ectotherms are thought to be especially vulnerable to climate change because most have a narrow range of thermal tolerance while living close to their upper thermal tolerance limits. Additionally, many tropical species live in closed-canopy forests, which provide homogenous thermal landscapes that prevent behavioral compensation for stressfully warm temperatures. Finally, tropical ectotherms are thought to have decreased capacity for phenotypic plasticity because they have evolved in thermally stable environments. -

Cell Cycle Arrest Through Indirect Transcriptional Repression by P53: I Have a DREAM
Cell Death and Differentiation (2018) 25, 114–132 Official journal of the Cell Death Differentiation Association OPEN www.nature.com/cdd Review Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM Kurt Engeland1 Activation of the p53 tumor suppressor can lead to cell cycle arrest. The key mechanism of p53-mediated arrest is transcriptional downregulation of many cell cycle genes. In recent years it has become evident that p53-dependent repression is controlled by the p53–p21–DREAM–E2F/CHR pathway (p53–DREAM pathway). DREAM is a transcriptional repressor that binds to E2F or CHR promoter sites. Gene regulation and deregulation by DREAM shares many mechanistic characteristics with the retinoblastoma pRB tumor suppressor that acts through E2F elements. However, because of its binding to E2F and CHR elements, DREAM regulates a larger set of target genes leading to regulatory functions distinct from pRB/E2F. The p53–DREAM pathway controls more than 250 mostly cell cycle-associated genes. The functional spectrum of these pathway targets spans from the G1 phase to the end of mitosis. Consequently, through downregulating the expression of gene products which are essential for progression through the cell cycle, the p53–DREAM pathway participates in the control of all checkpoints from DNA synthesis to cytokinesis including G1/S, G2/M and spindle assembly checkpoints. Therefore, defects in the p53–DREAM pathway contribute to a general loss of checkpoint control. Furthermore, deregulation of DREAM target genes promotes chromosomal instability and aneuploidy of cancer cells. Also, DREAM regulation is abrogated by the human papilloma virus HPV E7 protein linking the p53–DREAM pathway to carcinogenesis by HPV.Another feature of the pathway is that it downregulates many genes involved in DNA repair and telomere maintenance as well as Fanconi anemia. -

Epididymal V-Atpase-Rich Cell Proteome Database
A comprehensive list of the proteins that are expressed in V-ATPase-rich cells harvested from the epididymides based on the isolation by enzymatic digestion and fluorescence-activated cell sorting (FACS) from transgenic B1-EGFP mice, which express EGFP under the control of the promoter of the V-ATPase-B1 subunit. In these mice, narrow and clear cells of the epididymis express EGFP. The protein identification was performed by LC-MS/MS using an LTQ tandem mass spectrometer (Thermo Fisher Scientific). For questions or comments please contact Sylvie Breton ([email protected]) or Mark A. Knepper ([email protected]). -
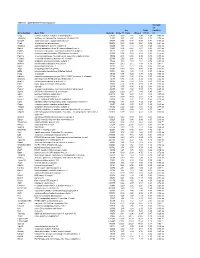
Table S-1 Mpkccd Transcriptome
Table S1. mpkCCD Cell Transcriptomes. Correlati on Ori. Ratio Coefficie Gene Symbol Gene Title GeneID Clone 11 Clone Clone 2 (11/2) nt Copg coatomer protein complex, subunit gamma 54161 1.99 1.81 2.48 0.80 -0.49 ns Atp6v0d1 ATPase, H+ transporting, lysosomal V0 subunit D1 11972 3.37 2.87 3.00 1.12 0.74 ns Golga7 golgi autoantigen, golgin subfamily a, 7 57437 3.03 3.40 3.58 0.84 -0.34 ns Psph phosphoserine phosphatase 100678 0.38 0.66 0.64 0.59 -0.61 ns Trappc4 trafficking protein particle complex 4 60409 1.17 1.12 1.08 1.08 0.64 ns Dpm2 dolichol-phosphate (beta-D) mannosyltransferase 2 13481 0.74 0.87 0.81 0.91 -0.31 ns Psmb5 proteasome (prosome, macropain) subunit, beta type 5 19173 2.92 3.00 2.92 0.99 0.32 ns Dhrs1 dehydrogenase/reductase (SDR family) member 1 52585 0.74 0.68 0.88 0.83 -0.18 ns Ppm1a protein phosphatase 1A, magnesium dependent, alpha isoform 19042 2.43 2.60 2.96 0.82 -0.53 ns Psenen presenilin enhancer 2 homolog (C. elegans) 66340 2.52 2.88 3.80 0.66 -0.77 ns Anapc1 anaphase promoting complex subunit 1 17222 1.16 1.33 1.61 0.72 -0.33 ns Mrpl43 mitochondrial ribosomal protein L43 94067 2.15 2.15 1.84 1.16 0.91 * Nmt1 N-myristoyltransferase 1 18107 3.21 2.71 3.36 0.95 0.31 ns Atg5 autophagy-related 5 (yeast) 11793 0.70 0.73 0.66 1.05 -0.23 ns Mtif2 mitochondrial translational initiation factor 2 76784 1.16 1.29 1.19 0.97 -0.36 ns Psap prosaposin 19156 8.07 7.25 8.37 0.96 0.02 ns Ube2g1 ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, C. -

1 Imipramine Treatment and Resiliency Exhibit Similar
Imipramine Treatment and Resiliency Exhibit Similar Chromatin Regulation in the Mouse Nucleus Accumbens in Depression Models Wilkinson et al. Supplemental Material 1. Supplemental Methods 2. Supplemental References for Tables 3. Supplemental Tables S1 – S24 SUPPLEMENTAL TABLE S1: Genes Demonstrating Increased Repressive DimethylK9/K27-H3 Methylation in the Social Defeat Model (p<0.001) SUPPLEMENTAL TABLE S2: Genes Demonstrating Decreased Repressive DimethylK9/K27-H3 Methylation in the Social Defeat Model (p<0.001) SUPPLEMENTAL TABLE S3: Genes Demonstrating Increased Repressive DimethylK9/K27-H3 Methylation in the Social Isolation Model (p<0.001) SUPPLEMENTAL TABLE S4: Genes Demonstrating Decreased Repressive DimethylK9/K27-H3 Methylation in the Social Isolation Model (p<0.001) SUPPLEMENTAL TABLE S5: Genes Demonstrating Common Altered Repressive DimethylK9/K27-H3 Methylation in the Social Defeat and Social Isolation Models (p<0.001) SUPPLEMENTAL TABLE S6: Genes Demonstrating Increased Repressive DimethylK9/K27-H3 Methylation in the Social Defeat and Social Isolation Models (p<0.001) SUPPLEMENTAL TABLE S7: Genes Demonstrating Decreased Repressive DimethylK9/K27-H3 Methylation in the Social Defeat and Social Isolation Models (p<0.001) SUPPLEMENTAL TABLE S8: Genes Demonstrating Increased Phospho-CREB Binding in the Social Defeat Model (p<0.001) SUPPLEMENTAL TABLE S9: Genes Demonstrating Decreased Phospho-CREB Binding in the Social Defeat Model (p<0.001) SUPPLEMENTAL TABLE S10: Genes Demonstrating Increased Phospho-CREB Binding in the Social -

Identification of Novel Substrates of Lrrk2, a Parkinson's Disease
IDENTIFICATION OF NOVEL SUBSTRATES OF LRRK2, A PARKINSON’S DISEASE ASSOCIATED KINASE By CAROLINE H. LEE Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Dissertation Adviser: Dr. Shu G. Chen Department of Biochemistry CASE WESTERN RESERVE UNIVERSITY May 2014 Case Western Reserve University School of Graduate Studies We hereby approve the thesis/dissertation of CAROLINE H. LEE Candidate for the degree of Doctor of Philosophy Committee Chair Dr. Michael E. Harris Committee Members Dr. Shu G. Chen Dr. Robert Petersen Dr. Hung-ying Kao Dr. Neena Singh Date of Defense December 18, 2013 *We also certify that written approval has been obtained for any proprietary material contained therein. 1 Table of Contents Table of Contents ...................................................................................................................... 2 List of Tables .............................................................................................................................. 4 List of Figures ............................................................................................................................. 5 List of Abbreviations ............................................................................................................... 6 Acknowledgements .................................................................................................................. 7 Abstract .................................................................................................................................... -

Examination of Kinase-Regulated Transcriptional Networks
Examinationof Kinase-RegulatedTranscriptional Networks Using Pharmacolosicaland ChemicalGenetic Tools by BeatriceT. Wanq DISSERTATION Submittedin partialsatisfaction of the requirementsfor the degreeof DOCTOROF PHILOSOPHY in Biochemistryand Molecular Biology in the GRADUATE DIVISION of the Copyright 2011 by Beatrice T. Wang ii Acknowledgements First and foremost, I thank my academic advisor Dr. Kevan Shokat for his guidance and support throughout my graduate career. Kevan has a gift for understanding complex biological systems and I was so fortunate to have had the opportunity to study under his mentorship. Even during difficult times, Kevan’s insight and perpetual optimism gave me the motivation I needed to succeed, and his admirable ability to achieve a work-life balance makes him an inspiring role model. I was also fortunate to have had the most kind and talented group of colleagues in the Shokat lab. In particular, I am indebted to Jasmina Allen for her dedicated mentorship that was crucial to my graduate learning experience, and to Morri Feldman for always generously offering me his time and ideas. Additionally, I appreciate Sasha Statsuk, Greg Ducker, Adam Garske, Ulf Peters, and Brandon Tavshanjian for not only technical advice but also insightful and entertaining conversations. I also thank Valerie Ohman for providing impeccable administrative assistance that the lab could not function without. I am also grateful to my thesis committee, Keith Yamamoto and Geeta Narlikar, for providing scientific support and career advice. I was supported in 2008-2009 by the University of California Cancer Research Coordinating Committee. Lastly, I would like to acknowledge my family, Ping, Teresa, and Clarice Wang for their love and support.