The Origin of a Derived Superkingdom: How a Gram-Positive Bacterium Crossed the Desert to Become an Archaeon
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Phagotrophic Origin of Eukaryotes and Phylogenetic Classification Of
International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 DOI: 10.1099/ijs.0.02058-0 The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa Department of Zoology, T. Cavalier-Smith University of Oxford, South Parks Road, Oxford OX1 3PS, UK Tel: j44 1865 281065. Fax: j44 1865 281310. e-mail: tom.cavalier-smith!zoo.ox.ac.uk Eukaryotes and archaebacteria form the clade neomura and are sisters, as shown decisively by genes fragmented only in archaebacteria and by many sequence trees. This sisterhood refutes all theories that eukaryotes originated by merging an archaebacterium and an α-proteobacterium, which also fail to account for numerous features shared specifically by eukaryotes and actinobacteria. I revise the phagotrophy theory of eukaryote origins by arguing that the essentially autogenous origins of most eukaryotic cell properties (phagotrophy, endomembrane system including peroxisomes, cytoskeleton, nucleus, mitosis and sex) partially overlapped and were synergistic with the symbiogenetic origin of mitochondria from an α-proteobacterium. These radical innovations occurred in a derivative of the neomuran common ancestor, which itself had evolved immediately prior to the divergence of eukaryotes and archaebacteria by drastic alterations to its eubacterial ancestor, an actinobacterial posibacterium able to make sterols, by replacing murein peptidoglycan by N-linked glycoproteins and a multitude of other shared neomuran novelties. The conversion of the rigid neomuran wall into a flexible surface coat and the associated origin of phagotrophy were instrumental in the evolution of the endomembrane system, cytoskeleton, nuclear organization and division and sexual life-cycles. Cilia evolved not by symbiogenesis but by autogenous specialization of the cytoskeleton. -
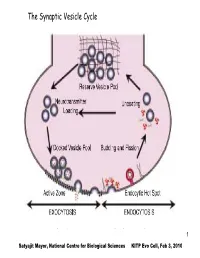
The Synaptic Vesicle Cycle
The Synaptic Vesicle Cycle 1 Satyajit Mayor, National Centre for Biological Sciences KITP Evo Cell, Feb 3, 2010 The Eukaryotic Commandments: T. Cavalier-Smith (i)origin of the endomembrane system (ER, Golgi and lysosomes) and coated- vesicle budding and fusion, including endocytosis and exocytosis; (ii) origin of the cytoskeleton, centrioles, cilia and associated molecular motors; (iii) origin of the nucleus, nuclear pore complex and trans-envelope protein and RNA transport; (iv) origin of linear chromosomes with plural replicons, centromeres and telomeres; (v) origin of novel cell-cycle controls and mitotic segregation; (vi) origin of sex (syngamy, nuclear fusion and meiosis); (vii) origin of peroxisomes; (ix) origin of mitochondria; (viii) novel patterns of rRNAprocessing using small nucleolar-ribonucleoproteins (snoRNPs); (x) origin of spliceosomal introns. 2 EM of Chemical synapse mitochondria Active Zone Figure 5.3, Bear, 2001 3 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. I phase: The ancestors of eukaryotes, the stem neomura, are shared with archaebacteria and evolved during the neomuran revolution, in which N-linked glycoproteins replaced murein peptidoglycan and 18 other suites of characters changed radically through adaptation of an ancestral actinobacterium to thermophily, as discussed in detail by Cavalier- Smith (2002a). 4 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. II phase: In the next phase, archaebacteria and eukaryotes diverged dramatically. Archaebacteria retained the wall and therefore their general bacterial cell and genetic organization, but became adapted to even hotter and more acidic environments by substituting prenyl ether lipids for the ancestral acyl esters and making new acid- resistant flagellar shafts (Cavalier-Smith, 2002a). -

Origin and Evolution of Eukaryotic Compartmentalization
TESI DOCTORAL UPF 2016 Origin and evolution of eukaryotic compartmentalization Alexandros Pittis Director Dr. Toni Gabaldon´ Comparative Genomics Group Bioinformatics and Genomics Department Centre for Genomic Regulation (CRG) To my father Stavros for the PhD he never did Acknowledgments Looking back at the years of my PhD in the Comparative Genomics group at CRG, I feel it was equally a process of scientific, as well as personal development. All this time I was very privileged to be surrounded by people that offered me their generous support in both fronts. And they were available in the very moments that I was fighting more myself than the unsolvable (anyway) comparative genomics puzzles. I hope that in the future I will have many times the chance to express them my appreciation, way beyond these few words. First, to Toni, my supervisor, for all his support, and patience, and confidence, and respect, especially at the moments that things did not seem that promising. He offered me freedom, to try, to think, to fail, to learn, to achieve that few enjoy during their PhD, and I am very thankful to him. Then, to my good friends and colleagues, those that I found in the group already, and others that joined after me. A very special thanks to Marinita and Jaime, for all their valuable time, and guidance and paradigm, which nevertheless I never managed to follow. They have both marked my phylogenomics path so far and I cannot escape. To Les and Salvi, my fellow students and pals at the time for all that we shared; to Gab and Fran and Dam, for -

Radical Views of the Tree of Life
Environmental Microbiology (2009) doi:10.1111/j.1462-2920.2009.01895.x Genomics update Radical views of the Tree of Life Howard Ochman* been posited over the years (Embley and Martin, 2006; Department of Biochemistry and Molecular Biophysics, Poole and Penny, 2007). University of Arizona, Tucson, Arizona 85718, USA. To examine the origin of the eukaryotes, Pisani et al. (2007) applied a supertree approach to each of three The tripartite division of cellular organisms into three large overlapping data sets containing either 0, 8 or 21 domains – the Bacteria, the Archaea and the Eukarya – eukaryotic genomes, in order to monitor the effects of has become so ingrained that most of us have all but taxon sampling on the resulting branching orders. Super- forgotten (or did not even know) that the ancestry and trees are phylogenies that are assembled by merging sets relationships among these ancient lineages are far from of more limited trees, each based on a gene that is not settled. The most widely accepted view, which emerged necessarily present in all taxa. Their results resolved bac- primarily from analyses of anciently duplicated genes teria and archaea as separate groups, but favour a chi- (Gogarten et al., 1989; Iwabe et al., 1989), places Bacte- meric origin of eukaryotes, as the majority of eukaryotic ria as the basal lineage and distinct from the common genes with prokaryotic homologues derive from multiple ancestor of archaea and eukaryotes. Accordingly, these symbioses. These findings help explain how different studies frame the Archaea, despite their appellation, as genes originated and why particular genes show distinct archaic but not primordial, and the three domains are phylogenetic histories, but they do not ascertain whether each viewed as separate lineages, with archaea and the cellular lineage leading to eukaryotes – i.e. -

Origin of the Cell Nucleus, Mitosis and Sex: Roles of Intracellular Coevolution Thomas Cavalier-Smith*
Cavalier-Smith Biology Direct 2010, 5:7 http://www.biology-direct.com/content/5/1/7 RESEARCH Open Access Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution Thomas Cavalier-Smith* Abstract Background: The transition from prokaryotes to eukaryotes was the most radical change in cell organisation since life began, with the largest ever burst of gene duplication and novelty. According to the coevolutionary theory of eukaryote origins, the fundamental innovations were the concerted origins of the endomembrane system and cytoskeleton, subsequently recruited to form the cell nucleus and coevolving mitotic apparatus, with numerous genetic eukaryotic novelties inevitable consequences of this compartmentation and novel DNA segregation mechanism. Physical and mutational mechanisms of origin of the nucleus are seldom considered beyond the long- standing assumption that it involved wrapping pre-existing endomembranes around chromatin. Discussions on the origin of sex typically overlook its association with protozoan entry into dormant walled cysts and the likely simultaneous coevolutionary, not sequential, origin of mitosis and meiosis. Results: I elucidate nuclear and mitotic coevolution, explaining the origins of dicer and small centromeric RNAs for positionally controlling centromeric heterochromatin, and how 27 major features of the cell nucleus evolved in four logical stages, making both mechanisms and selective advantages explicit: two initial stages (origin of 30 nm chromatin fibres, enabling DNA compaction; and firmer attachment of endomembranes to heterochromatin) protected DNA and nascent RNA from shearing by novel molecular motors mediating vesicle transport, division, and cytoplasmic motility. Then octagonal nuclear pore complexes (NPCs) arguably evolved from COPII coated vesicle proteins trapped in clumps by Ran GTPase-mediated cisternal fusion that generated the fenestrated nuclear envelope, preventing lethal complete cisternal fusion, and allowing passive protein and RNA exchange. -

Protists – a Textbook Example for a Paraphyletic Taxon
ARTICLE IN PRESS Organisms, Diversity & Evolution 7 (2007) 166–172 www.elsevier.de/ode Protists – A textbook example for a paraphyletic taxon$ Martin Schlegela,Ã, Norbert Hu¨lsmannb aInstitute for Biology II, University of Leipzig, Talstraße 33, 04103 Leipzig, Germany bFree University of Berlin, Institute of Biology/Zoology, Working group Protozoology, Ko¨nigin-Luise-Straße 1-3, 14195 Berlin, Germany Received 7 September 2004; accepted 21 November 2006 Abstract Protists constitute a paraphyletic taxon since the latter is based on the plesiomorphic character of unicellularity and does not contain all descendants of the stem species. Multicellularity evolved several times independently in metazoans, higher fungi, heterokonts, red and green algae. Various hypotheses have been developed on the evolution and nature of the eukaryotic cell, considering the accumulating data on the chimeric nature of the eukaryote genome. Subsequent evolution of the protists was further complicated by primary, secondary, and even tertiary intertaxonic recombinations. However, multi-gene sequence comparisons and structural data point to a managable number of such events. Several putative monophyletic lineages and a gross picture of eukaryote phylogeny are emerging on the basis of those data. The Chromalveolata comprise Chromista and Alveolata (Dinoflagellata, Apicomplexa, Ciliophora, Perkinsozoa, and Haplospora). Major lineages of the former ‘amoebae’ group within the Heterolobosa, Cercozoa, and Amoebozoa. Cercozoa, including filose testate amoebae, chlorarachnids, and plasmodiophoreans seem to be affiliated with foraminiferans. Amoebozoa consistently form the sister group of the Opisthokonta (including fungi, and with choanoflagellates as sister group of metazoans). A clade of ‘plants’ comprises glaucocystophytes, red algae, green algae, and land vascular plants. The controversial debate on the root of the eukaryote tree has been accelerated by the interpretation of gene fusions as apomorphic characters. -

The Gospel According to LUCA (The Last Universal Common Ancestor)
UC San Diego UC San Diego Electronic Theses and Dissertations Title The gospel according to LUCA (the last universal common ancestor) Permalink https://escholarship.org/uc/item/6pw6n8xp Author Valas, Ruben Eliezer Meyer Publication Date 2010 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO The gospel according to LUCA (the last universal common ancestor) A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Bioinformatics by Ruben Eliezer Meyer Valas Committee in charge: Professor Philip E. Bourne, Chair Professor William F. Loomis, Co-Chair Professor Russell F. Doolittle Professor Richard D. Norris Professor Milton H. Saier Jr. 2010 The Dissertation of Ruben Eliezer Meyer Valas is approved, and it is acceptable in quality and form for publication on microfilm and electronically: ______________________________________________________________________ ______________________________________________________________________ ______________________________________________________________________ ______________________________________________________________________ Co-chair ______________________________________________________________________ Chair University of California, San Diego 2010 iii DEDICATION I dedicate this work to Alexander “Sasha” Shulgin. Sasha is probably the wisest person I’ve met. He refuses to allow anything other than his own imagination dictates the rules of what is -

Genomic Analysis of Uncultured Microbes in Marine Sediments
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 12-2017 Singled Out: Genomic analysis of uncultured microbes in marine sediments Jordan Toby Bird University of Tennessee, [email protected] Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Recommended Citation Bird, Jordan Toby, "Singled Out: Genomic analysis of uncultured microbes in marine sediments. " PhD diss., University of Tennessee, 2017. https://trace.tennessee.edu/utk_graddiss/4829 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Jordan Toby Bird entitled "Singled Out: Genomic analysis of uncultured microbes in marine sediments." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the equirr ements for the degree of Doctor of Philosophy, with a major in Microbiology. Karen G. Lloyd, Major Professor We have read this dissertation and recommend its acceptance: Mircea Podar, Andrew D. Steen, Erik R. Zinser Accepted for the Council: Dixie L. Thompson Vice Provost and Dean of the Graduate School (Original signatures are on file with official studentecor r ds.) Singled Out: Genomic analysis of uncultured microbes in marine sediments A Dissertation Presented for the Doctor of Philosophy Degree The University of Tennessee, Knoxville Jordan Toby Bird December 2017 Copyright © 2017 by Jordan Bird All rights reserved. -

The Soil Microbial Food Web Revisited: Predatory Myxobacteria As Keystone Taxa?
The ISME Journal https://doi.org/10.1038/s41396-021-00958-2 ARTICLE The soil microbial food web revisited: Predatory myxobacteria as keystone taxa? 1 1 1,2 1 1 Sebastian Petters ● Verena Groß ● Andrea Söllinger ● Michelle Pichler ● Anne Reinhard ● 1 1 Mia Maria Bengtsson ● Tim Urich Received: 4 October 2018 / Revised: 24 February 2021 / Accepted: 4 March 2021 © The Author(s) 2021. This article is published with open access Abstract Trophic interactions are crucial for carbon cycling in food webs. Traditionally, eukaryotic micropredators are considered the major micropredators of bacteria in soils, although bacteria like myxobacteria and Bdellovibrio are also known bacterivores. Until recently, it was impossible to assess the abundance of prokaryotes and eukaryotes in soil food webs simultaneously. Using metatranscriptomic three-domain community profiling we identified pro- and eukaryotic micropredators in 11 European mineral and organic soils from different climes. Myxobacteria comprised 1.5–9.7% of all obtained SSU rRNA transcripts and more than 60% of all identified potential bacterivores in most soils. The name-giving and well-characterized fi 1234567890();,: 1234567890();,: predatory bacteria af liated with the Myxococcaceae were barely present, while Haliangiaceae and Polyangiaceae dominated. In predation assays, representatives of the latter showed prey spectra as broad as the Myxococcaceae. 18S rRNA transcripts from eukaryotic micropredators, like amoeba and nematodes, were generally less abundant than myxobacterial 16S rRNA transcripts, especially in mineral soils. Although SSU rRNA does not directly reflect organismic abundance, our findings indicate that myxobacteria could be keystone taxa in the soil microbial food web, with potential impact on prokaryotic community composition. -

The Evolutionary Tree of Life
This work by Garry Stevens is licensed under Life is easy. We know this because the Creative Commons licence CC-BY-NC-ND, earliest evidence dates from 3,800 million LUCA and is available for free from years ago (mya), shortly after the earth’s charts.archsoc.com. See terms of use there. seas stopped boiling, and complex Main source: Walker G., Dorrell R.G., chemical substances could survive without Schlacht A., Dacks J.B. (2011): Eukaryotic being ripped apart. NEOMURA Bacteria are "little bags of chemicals", EUBACTERIA systematics New walls The archaea and eubacteria are surrounded by a membrane. Usually they True bacteria collectively referred to as prokaryotes, since have either cilia (small hairs on the The cell walls of the Neomura do not contain neither have a membrane-enclosed The Tree of Life membrane surface) or flagella (a large a polymer called murein, which is present in nucleus. For almost half the history of life on bacteria. whip) to move around. They are typically 1% earth, they were the only living organisms. of the size of a eucaryote cell They reproduce by dividing in two. They are common pathogens of other organisms. Every Creature That Has Ever Lived ARCHAEA Archaea are as structurally simple as They are not pathogens, but instead are The eucarya are the product of an If the prokaryotes were easy to create, the EUCARYA True nucleus Old bacteria bacteria, but exhibit much more diverse important commensal or mutually beneficial archaeon engulfing a bacteria, and the next step to the eucaryotes was incredibly The eucaryotes have cells that contain chemical metabolisms; and can live in partners to many other organisms. -

Taxonomy (Biology)
Taxonomy (biology) In biology, taxonomy (from Ancient Greek τάξις (taxis) 'arrangement', and -νομία (- nomia) 'method') is the scientific study of naming, defining (circumscribing) and classifying groups of biological organisms based on shared characteristics. Organisms are grouped into taxa (singular: taxon) and these groups are given a taxonomic rank; groups of a given rank can be aggregated to form a super-group of higher rank, thus creating a taxonomic hierarchy. The principal ranks in modern use are domain, kingdom, phylum (division is sometimes used in botany in place of phylum), class, order, family, genus, and species. The Swedish botanist Carl Linnaeus is regarded as the founder of the current system of taxonomy, as he developed a system known as Linnaean taxonomy for categorizing organisms and binomial nomenclature for naming organisms. With the advent of such fields of study as phylogenetics, cladistics, and systematics, the Linnaean system has progressed to a system of modern biological classification based on the evolutionary relationships between organisms, both living and extinct. Definition The exact definition of taxonomy varies from source to source, but the core of the discipline remains: the conception, naming, and classification of groups of organisms.[1] As points of reference, recent definitions of taxonomy are presented below: 1. Theory and practice of grouping individuals into species, arranging species into larger groups, and giving those groups names, thus producing a classification.[2] 2. A field of science (and major component of systematics) that encompasses description, identification, nomenclature, and classification[3] 3. The science of classification, in biology the arrangement of organisms into a classification[4] 4. -

Reconciling an Archaeal Origin of Eukaryotes with Engulfment: a Biologically Plausible Update of the Eocyte Hypothesis
Research in Microbiology 162 (2011) 71e76 www.elsevier.com/locate/resmic Reconciling an archaeal origin of eukaryotes with engulfment: a biologically plausible update of the Eocyte hypothesis Anthony M. Poole a,b,*, Nadja Neumann a a Department of Molecular Biology and Functional Genomics, Stockholm University, SE-106 91 Stockholm, Sweden b School of Biological Sciences, University of Canterbury, Private Bag 4800, Christchurch 8140, New Zealand Available online 27 October 2010 Abstract An archaeal origin of eukaryotes is often equated with the engulfment of the bacterial ancestor of mitochondria by an archaeon. Such an event is problematic in that it is not supported by archaeal cell biology. We show that placing phylogenetic results within a stem-and-crown framework eliminates such incompatibilities, and that an archaeal origin for eukaryotes (as suggested from recent phylogenies) can be uncontroversially reconciled with phagocytosis as the mechanism for engulfment of the mitochondrial ancestor. This is significant because it eliminates a perceived problem with eukaryote origins: that an archaeal origin of eukaryotes (as under the Eocyte hypothesis) cannot be reconciled with existing cell biological mechanisms through which bacteria may take up residence inside eukaryote cells. Ó 2010 Institut Pasteur. Published by Elsevier Masson SAS. All rights reserved. Keywords: Stem group; Crown group; Eocyte hypothesis; Phagocytosis; Eukaryotes; Archaea 1. Introduction derived. This also appears to be the case for key parts of the machinery for phagocytosisdcell engulfment (Yutin et al., A range of models for the origin of the eukaryote cell have 2009). As summarised in Table 1, the emerging consensus been proposed on phylogenetic, genomic and cell biological from a range of studies is that the LECA was essentially grounds (reviewed in Embley and Martin, 2006; Martin et al., a fully-fledged eukaryote cell.