A Formal Analysis of Phylogenetic Terminology: Towards a Reconsideration of the Current Paradigm in Systematics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Species Concepts Should Not Conflict with Evolutionary History, but Often Do
ARTICLE IN PRESS Stud. Hist. Phil. Biol. & Biomed. Sci. xxx (2008) xxx–xxx Contents lists available at ScienceDirect Stud. Hist. Phil. Biol. & Biomed. Sci. journal homepage: www.elsevier.com/locate/shpsc Species concepts should not conflict with evolutionary history, but often do Joel D. Velasco Department of Philosophy, University of Wisconsin-Madison, 5185 White Hall, 600 North Park St., Madison, WI 53719, USA Department of Philosophy, Building 90, Stanford University, Stanford, CA 94305, USA article info abstract Keywords: Many phylogenetic systematists have criticized the Biological Species Concept (BSC) because it distorts Biological Species Concept evolutionary history. While defences against this particular criticism have been attempted, I argue that Phylogenetic Species Concept these responses are unsuccessful. In addition, I argue that the source of this problem leads to previously Phylogenetic Trees unappreciated, and deeper, fatal objections. These objections to the BSC also straightforwardly apply to Taxonomy other species concepts that are not defined by genealogical history. What is missing from many previous discussions is the fact that the Tree of Life, which represents phylogenetic history, is independent of our choice of species concept. Some species concepts are consistent with species having unique positions on the Tree while others, including the BSC, are not. Since representing history is of primary importance in evolutionary biology, these problems lead to the conclusion that the BSC, along with many other species concepts, are unacceptable. If species are to be taxa used in phylogenetic inferences, we need a history- based species concept. Ó 2008 Elsevier Ltd. All rights reserved. When citing this paper, please use the full journal title Studies in History and Philosophy of Biological and Biomedical Sciences 1. -

Reticulate Evolution and Phylogenetic Networks 6
CHAPTER Reticulate Evolution and Phylogenetic Networks 6 So far, we have modeled the evolutionary process as a bifurcating treelike process. The main feature of trees is that the flow of information in one lineage is independent of that in another lineage. In real life, however, many process- es—including recombination, hybridization, polyploidy, introgression, genome fusion, endosymbiosis, and horizontal gene transfer—do not abide by lineage independence and cannot be modeled by trees. Reticulate evolution refers to the origination of lineages through the com- plete or partial merging of two or more ancestor lineages. The nonvertical con- nections between lineages are referred to as reticulations. In this chapter, we introduce the language of networks and explain how networks can be used to study non-treelike phylogenetic processes. A substantial part of this chapter will be dedicated to the evolutionary relationships among prokaryotes and the very intriguing question of the origin of the eukaryotic cell. Networks Networks were briefly introduced in Chapter 4 in the context of protein-protein interactions. Here, we provide a more formal description of networks and their use in phylogenetics. As mentioned in Chapter 5, trees are connected graphs in which any two nodes are connected by a single path. In phylogenetics, a network is a connected graph in which at least two nodes are connected by two or more pathways. In other words, a network is a connected graph that is not a tree. All networks contain at least one cycle, i.e., a path of branches that can be followed in one direction from any starting point back to the starting point. -

Phylogenetic Trees and Cladograms Are Graphical Representations (Models) of Evolutionary History That Can Be Tested
AP Biology Lab/Cladograms and Phylogenetic Trees Name _______________________________ Relationship to the AP Biology Curriculum Framework Big Idea 1: The process of evolution drives the diversity and unity of life. Essential knowledge 1.B.2: Phylogenetic trees and cladograms are graphical representations (models) of evolutionary history that can be tested. Learning Objectives: LO 1.17 The student is able to pose scientific questions about a group of organisms whose relatedness is described by a phylogenetic tree or cladogram in order to (1) identify shared characteristics, (2) make inferences about the evolutionary history of the group, and (3) identify character data that could extend or improve the phylogenetic tree. LO 1.18 The student is able to evaluate evidence provided by a data set in conjunction with a phylogenetic tree or a simple cladogram to determine evolutionary history and speciation. LO 1.19 The student is able create a phylogenetic tree or simple cladogram that correctly represents evolutionary history and speciation from a provided data set. [Introduction] Cladistics is the study of evolutionary classification. Cladograms show evolutionary relationships among organisms. Comparative morphology investigates characteristics for homology and analogy to determine which organisms share a recent common ancestor. A cladogram will begin by grouping organisms based on a characteristics displayed by ALL the members of the group. Subsequently, the larger group will contain increasingly smaller groups that share the traits of the groups before them. However, they also exhibit distinct changes as the new species evolve. Further, molecular evidence from genes which rarely mutate can provide molecular clocks that tell us how long ago organisms diverged, unlocking the secrets of organisms that may have similar convergent morphology but do not share a recent common ancestor. -

An Introduction to Phylogenetic Analysis
This article reprinted from: Kosinski, R.J. 2006. An introduction to phylogenetic analysis. Pages 57-106, in Tested Studies for Laboratory Teaching, Volume 27 (M.A. O'Donnell, Editor). Proceedings of the 27th Workshop/Conference of the Association for Biology Laboratory Education (ABLE), 383 pages. Compilation copyright © 2006 by the Association for Biology Laboratory Education (ABLE) ISBN 1-890444-09-X All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording, or otherwise, without the prior written permission of the copyright owner. Use solely at one’s own institution with no intent for profit is excluded from the preceding copyright restriction, unless otherwise noted on the copyright notice of the individual chapter in this volume. Proper credit to this publication must be included in your laboratory outline for each use; a sample citation is given above. Upon obtaining permission or with the “sole use at one’s own institution” exclusion, ABLE strongly encourages individuals to use the exercises in this proceedings volume in their teaching program. Although the laboratory exercises in this proceedings volume have been tested and due consideration has been given to safety, individuals performing these exercises must assume all responsibilities for risk. The Association for Biology Laboratory Education (ABLE) disclaims any liability with regards to safety in connection with the use of the exercises in this volume. The focus of ABLE is to improve the undergraduate biology laboratory experience by promoting the development and dissemination of interesting, innovative, and reliable laboratory exercises. -

Splits and Phylogenetic Networks
SplitsSplits andand PhylogeneticPhylogenetic NetworksNetworks DanielDaniel H.H. HusonHuson Paris, June 21, 2005 1 ContentsContents 1.1. PhylogeneticPhylogenetic treestrees 2.2. SplitsSplits networksnetworks 3.3. ConsensusConsensus networksnetworks 4.4. HybridizationHybridization andand reticulatereticulate networksnetworks 5.5. RecombinationRecombination networksnetworks 2 PhylogeneticPhylogenetic NetworksNetworks Bandelt (1991): Network displaying evolutionary relationships Other types of Splits Phylogenetic Reticulate phylogenetic networks trees networks networks Median Consensus Hybridization Recombination Augmented networks (super) networks networks trees networks Special case: “Galled trees” Any graph Ancestor Split decomposition, representing Split decomposition, recombination evolutionary Neighbor-net graphs data 3 PhylogeneticPhylogenetic NetworksNetworks 22 11 Other types of Splits Phylogenetic Reticulate phylogenetic networks trees networks networks 33 44 55 Median Consensus Hybridization Recombination Augmented networks (super) networks networks trees networks from sequences Special case: “Galled trees” from trees Any graph Ancestor Split decomposition, representing Split decomposition, recombination evolutionary Neighbor-net graphs data from distances 4 PhylogeneticPhylogenetic NetworksNetworksDan Gusfield: “Phylogenetic network” 22 11 “Generalized Other types of Splits Phylogenetic Reticulate phylogeneticphylogenetic networks trees networks network”networks 33 44 \ 55 Median Consensus Hybridization Recombination“More generalizedAugmented -

The Phagotrophic Origin of Eukaryotes and Phylogenetic Classification Of
International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 DOI: 10.1099/ijs.0.02058-0 The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa Department of Zoology, T. Cavalier-Smith University of Oxford, South Parks Road, Oxford OX1 3PS, UK Tel: j44 1865 281065. Fax: j44 1865 281310. e-mail: tom.cavalier-smith!zoo.ox.ac.uk Eukaryotes and archaebacteria form the clade neomura and are sisters, as shown decisively by genes fragmented only in archaebacteria and by many sequence trees. This sisterhood refutes all theories that eukaryotes originated by merging an archaebacterium and an α-proteobacterium, which also fail to account for numerous features shared specifically by eukaryotes and actinobacteria. I revise the phagotrophy theory of eukaryote origins by arguing that the essentially autogenous origins of most eukaryotic cell properties (phagotrophy, endomembrane system including peroxisomes, cytoskeleton, nucleus, mitosis and sex) partially overlapped and were synergistic with the symbiogenetic origin of mitochondria from an α-proteobacterium. These radical innovations occurred in a derivative of the neomuran common ancestor, which itself had evolved immediately prior to the divergence of eukaryotes and archaebacteria by drastic alterations to its eubacterial ancestor, an actinobacterial posibacterium able to make sterols, by replacing murein peptidoglycan by N-linked glycoproteins and a multitude of other shared neomuran novelties. The conversion of the rigid neomuran wall into a flexible surface coat and the associated origin of phagotrophy were instrumental in the evolution of the endomembrane system, cytoskeleton, nuclear organization and division and sexual life-cycles. Cilia evolved not by symbiogenesis but by autogenous specialization of the cytoskeleton. -
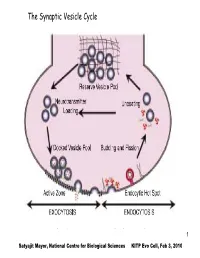
The Synaptic Vesicle Cycle
The Synaptic Vesicle Cycle 1 Satyajit Mayor, National Centre for Biological Sciences KITP Evo Cell, Feb 3, 2010 The Eukaryotic Commandments: T. Cavalier-Smith (i)origin of the endomembrane system (ER, Golgi and lysosomes) and coated- vesicle budding and fusion, including endocytosis and exocytosis; (ii) origin of the cytoskeleton, centrioles, cilia and associated molecular motors; (iii) origin of the nucleus, nuclear pore complex and trans-envelope protein and RNA transport; (iv) origin of linear chromosomes with plural replicons, centromeres and telomeres; (v) origin of novel cell-cycle controls and mitotic segregation; (vi) origin of sex (syngamy, nuclear fusion and meiosis); (vii) origin of peroxisomes; (ix) origin of mitochondria; (viii) novel patterns of rRNAprocessing using small nucleolar-ribonucleoproteins (snoRNPs); (x) origin of spliceosomal introns. 2 EM of Chemical synapse mitochondria Active Zone Figure 5.3, Bear, 2001 3 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. I phase: The ancestors of eukaryotes, the stem neomura, are shared with archaebacteria and evolved during the neomuran revolution, in which N-linked glycoproteins replaced murein peptidoglycan and 18 other suites of characters changed radically through adaptation of an ancestral actinobacterium to thermophily, as discussed in detail by Cavalier- Smith (2002a). 4 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. II phase: In the next phase, archaebacteria and eukaryotes diverged dramatically. Archaebacteria retained the wall and therefore their general bacterial cell and genetic organization, but became adapted to even hotter and more acidic environments by substituting prenyl ether lipids for the ancestral acyl esters and making new acid- resistant flagellar shafts (Cavalier-Smith, 2002a). -

Interpreting Cladograms Notes
Interpreting Cladograms Notes INTERPRETING CLADOGRAMS BIG IDEA: PHYLOGENIES DEPICT ANCESTOR AND DESCENDENT RELATIONSHIPS AMONG ORGANISMS BASED ON HOMOLOGY THESE EVOLUTIONARY RELATIONSHIPS ARE REPRESENTED BY DIAGRAMS CALLED CLADOGRAMS (BRANCHING DIAGRAMS THAT ORGANIZE RELATIONSHIPS) What is a Cladogram ● A diagram which shows ● Is not an evolutionary ___________ among tree, ___________show organisms. how ancestors are related ● Lines branching off other to descendants or how lines. The lines can be much they have changed traced back to where they branch off. These branching off points represent a hypothetical ancestor. Parts of a Cladogram Reading Cladograms ● Read like a family tree: show ________of shared ancestry between lineages. • When an ancestral lineage______: speciation is indicated due to the “arrival” of some new trait. Each lineage has unique ____to itself alone and traits that are shared with other lineages. each lineage has _______that are unique to that lineage and ancestors that are shared with other lineages — common ancestors. Quick Question #1 ●What is a_________? ● A group that includes a common ancestor and all the descendants (living and extinct) of that ancestor. Reading Cladogram: Identifying Clades ● Using a cladogram, it is easy to tell if a group of lineages forms a clade. ● Imagine clipping a single branch off the phylogeny ● all of the organisms on that pruned branch make up a clade Quick Question #2 ● Looking at the image to the right: ● Is the green box a clade? ● The blue? ● The pink? ● The orange? Reading Cladograms: Clades ● Clades are nested within one another ● they form a nested hierarchy. ● A clade may include many thousands of species or just a few. -

Conceptual Issues in Phylogeny, Taxonomy, and Nomenclature
Contributions to Zoology, 66 (1) 3-41 (1996) SPB Academic Publishing bv, Amsterdam Conceptual issues in phylogeny, taxonomy, and nomenclature Alexandr P. Rasnitsyn Paleontological Institute, Russian Academy ofSciences, Profsoyuznaya Street 123, J17647 Moscow, Russia Keywords: Phylogeny, taxonomy, phenetics, cladistics, phylistics, principles of nomenclature, type concept, paleoentomology, Xyelidae (Vespida) Abstract On compare les trois approches taxonomiques principales développées jusqu’à présent, à savoir la phénétique, la cladis- tique et la phylistique (= systématique évolutionnaire). Ce Phylogenetic hypotheses are designed and tested (usually in dernier terme s’applique à une approche qui essaie de manière implicit form) on the basis ofa set ofpresumptions, that is, of à les traits fondamentaux de la taxonomic statements explicite représenter describing a certain order of things in nature. These traditionnelle en de leur et particulier son usage preuves ayant statements are to be accepted as such, no matter whatever source en même temps dans la similitude et dans les relations de evidence for them exists, but only in the absence ofreasonably parenté des taxons en question. L’approche phylistique pré- sound evidence pleading against them. A set ofthe most current sente certains avantages dans la recherche de réponses aux phylogenetic presumptions is discussed, and a factual example problèmes fondamentaux de la taxonomie. ofa practical realization of the approach is presented. L’auteur considère la nomenclature A is made of the three -

Chapter 7 Phylogenesis Versus
108 The Evolution of Culturol Diversity Clades Lineages CHAPTER 7 PHYLOGENESIS VERSUS ETHNOGENESIS IN vvvv TURKMEN CULTURAL EVOLUTION vv v Mark Collard and Jamshid Tehrani INTRODUCTION The processes responsible for producing the similarities and differences among cultures have been the focus of much debate in recent years, as has the corollary vv issue of linking cultural data with the patterns recorded by linguists and Figure 6.10 Clodes versus lineages. All nine diagrams represent the same hiologists working with human populations (eg, Romney 1957; Vogt 1964; phylogeny, with clades highlighted on the left and lineages on the tight Chakraborty ct a11976; Brace and Hinton 1981; Cavalli-Sforza and Feldman 1981; Additional lineages can be counted from various internal nodes to the branch Lumsden and Wilson 1981~ Ammerman and Cavalli-Sforza 1984; Boyd and tips (after de Quelroz 1998). Richerson 1985; Terrell 1986, 1988; Kirch and Green 1987, 2001; Renfrew 19H7, 1992, 2000b, 2001; Atkinson 1989; Croes 1989; Bateman et a11990; Durham 1990, Archaeologists are uniquely capable of ans\vering these questions, and cladistics 1991,1992; Moore 1994b; Cavalli-Sforza and Cavalli-Sforza 1995; Guglielmino et a[ offers a means to answer them. 19Q5; Laland et af 1995; Zvelebil 1995; Bellwood 1996a, 2001; Boyd clal 1997~ But are we simply borrowing techniques of biological origin \vithout a firm Shennan 2000, 2002; Smith 2001; Whaley 2001; Terrell cI ill 20CH; Jordan dnd basis for so doing? No. We view cultural phenomena as residing in a series of Shennan 2003). To date, this debate has concentrated on two cornpeting nested hierarchies that comprise traditions, or lineages, at ever more-inclusive hypotheses, which have been termed the 'genetic', 'demie diffusion', 'branching' scales and that are held together by cultural as \veU as genetic transmission. -

Origin of the Cell Nucleus, Mitosis and Sex: Roles of Intracellular Coevolution Thomas Cavalier-Smith*
Cavalier-Smith Biology Direct 2010, 5:7 http://www.biology-direct.com/content/5/1/7 RESEARCH Open Access Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution Thomas Cavalier-Smith* Abstract Background: The transition from prokaryotes to eukaryotes was the most radical change in cell organisation since life began, with the largest ever burst of gene duplication and novelty. According to the coevolutionary theory of eukaryote origins, the fundamental innovations were the concerted origins of the endomembrane system and cytoskeleton, subsequently recruited to form the cell nucleus and coevolving mitotic apparatus, with numerous genetic eukaryotic novelties inevitable consequences of this compartmentation and novel DNA segregation mechanism. Physical and mutational mechanisms of origin of the nucleus are seldom considered beyond the long- standing assumption that it involved wrapping pre-existing endomembranes around chromatin. Discussions on the origin of sex typically overlook its association with protozoan entry into dormant walled cysts and the likely simultaneous coevolutionary, not sequential, origin of mitosis and meiosis. Results: I elucidate nuclear and mitotic coevolution, explaining the origins of dicer and small centromeric RNAs for positionally controlling centromeric heterochromatin, and how 27 major features of the cell nucleus evolved in four logical stages, making both mechanisms and selective advantages explicit: two initial stages (origin of 30 nm chromatin fibres, enabling DNA compaction; and firmer attachment of endomembranes to heterochromatin) protected DNA and nascent RNA from shearing by novel molecular motors mediating vesicle transport, division, and cytoplasmic motility. Then octagonal nuclear pore complexes (NPCs) arguably evolved from COPII coated vesicle proteins trapped in clumps by Ran GTPase-mediated cisternal fusion that generated the fenestrated nuclear envelope, preventing lethal complete cisternal fusion, and allowing passive protein and RNA exchange. -

Protists – a Textbook Example for a Paraphyletic Taxon
ARTICLE IN PRESS Organisms, Diversity & Evolution 7 (2007) 166–172 www.elsevier.de/ode Protists – A textbook example for a paraphyletic taxon$ Martin Schlegela,Ã, Norbert Hu¨lsmannb aInstitute for Biology II, University of Leipzig, Talstraße 33, 04103 Leipzig, Germany bFree University of Berlin, Institute of Biology/Zoology, Working group Protozoology, Ko¨nigin-Luise-Straße 1-3, 14195 Berlin, Germany Received 7 September 2004; accepted 21 November 2006 Abstract Protists constitute a paraphyletic taxon since the latter is based on the plesiomorphic character of unicellularity and does not contain all descendants of the stem species. Multicellularity evolved several times independently in metazoans, higher fungi, heterokonts, red and green algae. Various hypotheses have been developed on the evolution and nature of the eukaryotic cell, considering the accumulating data on the chimeric nature of the eukaryote genome. Subsequent evolution of the protists was further complicated by primary, secondary, and even tertiary intertaxonic recombinations. However, multi-gene sequence comparisons and structural data point to a managable number of such events. Several putative monophyletic lineages and a gross picture of eukaryote phylogeny are emerging on the basis of those data. The Chromalveolata comprise Chromista and Alveolata (Dinoflagellata, Apicomplexa, Ciliophora, Perkinsozoa, and Haplospora). Major lineages of the former ‘amoebae’ group within the Heterolobosa, Cercozoa, and Amoebozoa. Cercozoa, including filose testate amoebae, chlorarachnids, and plasmodiophoreans seem to be affiliated with foraminiferans. Amoebozoa consistently form the sister group of the Opisthokonta (including fungi, and with choanoflagellates as sister group of metazoans). A clade of ‘plants’ comprises glaucocystophytes, red algae, green algae, and land vascular plants. The controversial debate on the root of the eukaryote tree has been accelerated by the interpretation of gene fusions as apomorphic characters.