Protists – a Textbook Example for a Paraphyletic Taxon
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of a Novel Fused Gene Family Implicates Convergent
Chen et al. BMC Genomics (2018) 19:306 https://doi.org/10.1186/s12864-018-4685-y RESEARCH ARTICLE Open Access Identification of a novel fused gene family implicates convergent evolution in eukaryotic calcium signaling Fei Chen1,2,3, Liangsheng Zhang1, Zhenguo Lin4 and Zong-Ming Max Cheng2,3* Abstract Background: Both calcium signals and protein phosphorylation responses are universal signals in eukaryotic cell signaling. Currently three pathways have been characterized in different eukaryotes converting the Ca2+ signals to the protein phosphorylation responses. All these pathways have based mostly on studies in plants and animals. Results: Based on the exploration of genomes and transcriptomes from all the six eukaryotic supergroups, we report here in Metakinetoplastina protists a novel gene family. This family, with a proposed name SCAMK,comprisesSnRK3 fused calmodulin-like III kinase genes and was likely evolved through the insertion of a calmodulin-like3 gene into an SnRK3 gene by unequal crossover of homologous chromosomes in meiosis cell. Its origin dated back to the time intersection at least 450 million-year-ago when Excavata parasites, Vertebrata hosts, and Insecta vectors evolved. We also analyzed SCAMK’s unique expression pattern and structure, and proposed it as one of the leading calcium signal conversion pathways in Excavata parasite. These characters made SCAMK gene as a potential drug target for treating human African trypanosomiasis. Conclusions: This report identified a novel gene fusion and dated its precise fusion time -

Ptolemeba N. Gen., a Novel Genus of Hartmannellid Amoebae (Tubulinea, Amoebozoa); with an Emphasis on the Taxonomy of Saccamoeba
The Journal of Published by the International Society of Eukaryotic Microbiology Protistologists Journal of Eukaryotic Microbiology ISSN 1066-5234 ORIGINAL ARTICLE Ptolemeba n. gen., a Novel Genus of Hartmannellid Amoebae (Tubulinea, Amoebozoa); with an Emphasis on the Taxonomy of Saccamoeba Pamela M. Watsona, Stephanie C. Sorrella & Matthew W. Browna,b a Department of Biological Sciences, Mississippi State University, Mississippi State, Mississippi, 39762 b Institute for Genomics, Biocomputing & Biotechnology, Mississippi State University, Mississippi State, Mississippi, 39762 Keywords ABSTRACT 18S rRNA; amoeba; amoeboid; Cashia; cristae; freshwater amoebae; Hartmannella; Hartmannellid amoebae are an unnatural assemblage of amoeboid organisms mitochondrial morphology; SSU rDNA; SSU that are morphologically difficult to discern from one another. In molecular phy- rRNA; terrestrial amoebae; tubulinid. logenetic trees of the nuclear-encoded small subunit rDNA, they occupy at least five lineages within Tubulinea, a well-supported clade in Amoebozoa. The Correspondence polyphyletic nature of the hartmannellids has led to many taxonomic problems, M.W. Brown, Department of Biological in particular paraphyletic genera. Recent taxonomic revisions have alleviated Sciences, Mississippi State University, some of the problems. However, the genus Saccamoeba is paraphyletic and is Mississippi State, MS 39762, USA still in need of revision as it currently occupies two distinct lineages. Here, we Telephone number: +1 662-325-2406; report a new clade on the tree of Tubulinea, which we infer represents a novel FAX number: +1 662-325-7939; genus that we name Ptolemeba n. gen. This genus subsumes a clade of hart- e-mail: [email protected] mannellid amoebae that were previously considered in the genus Saccamoeba, but whose mitochondrial morphology is distinct from Saccamoeba. -

Number of Living Species in Australia and the World
Numbers of Living Species in Australia and the World 2nd edition Arthur D. Chapman Australian Biodiversity Information Services australia’s nature Toowoomba, Australia there is more still to be discovered… Report for the Australian Biological Resources Study Canberra, Australia September 2009 CONTENTS Foreword 1 Insecta (insects) 23 Plants 43 Viruses 59 Arachnida Magnoliophyta (flowering plants) 43 Protoctista (mainly Introduction 2 (spiders, scorpions, etc) 26 Gymnosperms (Coniferophyta, Protozoa—others included Executive Summary 6 Pycnogonida (sea spiders) 28 Cycadophyta, Gnetophyta under fungi, algae, Myriapoda and Ginkgophyta) 45 Chromista, etc) 60 Detailed discussion by Group 12 (millipedes, centipedes) 29 Ferns and Allies 46 Chordates 13 Acknowledgements 63 Crustacea (crabs, lobsters, etc) 31 Bryophyta Mammalia (mammals) 13 Onychophora (velvet worms) 32 (mosses, liverworts, hornworts) 47 References 66 Aves (birds) 14 Hexapoda (proturans, springtails) 33 Plant Algae (including green Reptilia (reptiles) 15 Mollusca (molluscs, shellfish) 34 algae, red algae, glaucophytes) 49 Amphibia (frogs, etc) 16 Annelida (segmented worms) 35 Fungi 51 Pisces (fishes including Nematoda Fungi (excluding taxa Chondrichthyes and (nematodes, roundworms) 36 treated under Chromista Osteichthyes) 17 and Protoctista) 51 Acanthocephala Agnatha (hagfish, (thorny-headed worms) 37 Lichen-forming fungi 53 lampreys, slime eels) 18 Platyhelminthes (flat worms) 38 Others 54 Cephalochordata (lancelets) 19 Cnidaria (jellyfish, Prokaryota (Bacteria Tunicata or Urochordata sea anenomes, corals) 39 [Monera] of previous report) 54 (sea squirts, doliolids, salps) 20 Porifera (sponges) 40 Cyanophyta (Cyanobacteria) 55 Invertebrates 21 Other Invertebrates 41 Chromista (including some Hemichordata (hemichordates) 21 species previously included Echinodermata (starfish, under either algae or fungi) 56 sea cucumbers, etc) 22 FOREWORD In Australia and around the world, biodiversity is under huge Harnessing core science and knowledge bases, like and growing pressure. -

The Phagotrophic Origin of Eukaryotes and Phylogenetic Classification Of
International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 DOI: 10.1099/ijs.0.02058-0 The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa Department of Zoology, T. Cavalier-Smith University of Oxford, South Parks Road, Oxford OX1 3PS, UK Tel: j44 1865 281065. Fax: j44 1865 281310. e-mail: tom.cavalier-smith!zoo.ox.ac.uk Eukaryotes and archaebacteria form the clade neomura and are sisters, as shown decisively by genes fragmented only in archaebacteria and by many sequence trees. This sisterhood refutes all theories that eukaryotes originated by merging an archaebacterium and an α-proteobacterium, which also fail to account for numerous features shared specifically by eukaryotes and actinobacteria. I revise the phagotrophy theory of eukaryote origins by arguing that the essentially autogenous origins of most eukaryotic cell properties (phagotrophy, endomembrane system including peroxisomes, cytoskeleton, nucleus, mitosis and sex) partially overlapped and were synergistic with the symbiogenetic origin of mitochondria from an α-proteobacterium. These radical innovations occurred in a derivative of the neomuran common ancestor, which itself had evolved immediately prior to the divergence of eukaryotes and archaebacteria by drastic alterations to its eubacterial ancestor, an actinobacterial posibacterium able to make sterols, by replacing murein peptidoglycan by N-linked glycoproteins and a multitude of other shared neomuran novelties. The conversion of the rigid neomuran wall into a flexible surface coat and the associated origin of phagotrophy were instrumental in the evolution of the endomembrane system, cytoskeleton, nuclear organization and division and sexual life-cycles. Cilia evolved not by symbiogenesis but by autogenous specialization of the cytoskeleton. -

A Revised Classification of Naked Lobose Amoebae (Amoebozoa
Protist, Vol. 162, 545–570, October 2011 http://www.elsevier.de/protis Published online date 28 July 2011 PROTIST NEWS A Revised Classification of Naked Lobose Amoebae (Amoebozoa: Lobosa) Introduction together constitute the amoebozoan subphy- lum Lobosa, which never have cilia or flagella, Molecular evidence and an associated reevaluation whereas Variosea (as here revised) together with of morphology have recently considerably revised Mycetozoa and Archamoebea are now grouped our views on relationships among the higher-level as the subphylum Conosa, whose constituent groups of amoebae. First of all, establishing the lineages either have cilia or flagella or have lost phylum Amoebozoa grouped all lobose amoe- them secondarily (Cavalier-Smith 1998, 2009). boid protists, whether naked or testate, aerobic Figure 1 is a schematic tree showing amoebozoan or anaerobic, with the Mycetozoa and Archamoe- relationships deduced from both morphology and bea (Cavalier-Smith 1998), and separated them DNA sequences. from both the heterolobosean amoebae (Page and The first attempt to construct a congruent molec- Blanton 1985), now belonging in the phylum Per- ular and morphological system of Amoebozoa by colozoa - Cavalier-Smith and Nikolaev (2008), and Cavalier-Smith et al. (2004) was limited by the the filose amoebae that belong in other phyla lack of molecular data for many amoeboid taxa, (notably Cercozoa: Bass et al. 2009a; Howe et al. which were therefore classified solely on morpho- 2011). logical evidence. Smirnov et al. (2005) suggested The phylum Amoebozoa consists of naked and another system for naked lobose amoebae only; testate lobose amoebae (e.g. Amoeba, Vannella, this left taxa with no molecular data incertae sedis, Hartmannella, Acanthamoeba, Arcella, Difflugia), which limited its utility. -
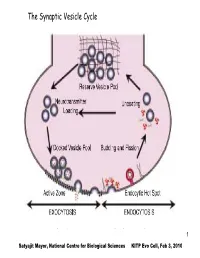
The Synaptic Vesicle Cycle
The Synaptic Vesicle Cycle 1 Satyajit Mayor, National Centre for Biological Sciences KITP Evo Cell, Feb 3, 2010 The Eukaryotic Commandments: T. Cavalier-Smith (i)origin of the endomembrane system (ER, Golgi and lysosomes) and coated- vesicle budding and fusion, including endocytosis and exocytosis; (ii) origin of the cytoskeleton, centrioles, cilia and associated molecular motors; (iii) origin of the nucleus, nuclear pore complex and trans-envelope protein and RNA transport; (iv) origin of linear chromosomes with plural replicons, centromeres and telomeres; (v) origin of novel cell-cycle controls and mitotic segregation; (vi) origin of sex (syngamy, nuclear fusion and meiosis); (vii) origin of peroxisomes; (ix) origin of mitochondria; (viii) novel patterns of rRNAprocessing using small nucleolar-ribonucleoproteins (snoRNPs); (x) origin of spliceosomal introns. 2 EM of Chemical synapse mitochondria Active Zone Figure 5.3, Bear, 2001 3 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. I phase: The ancestors of eukaryotes, the stem neomura, are shared with archaebacteria and evolved during the neomuran revolution, in which N-linked glycoproteins replaced murein peptidoglycan and 18 other suites of characters changed radically through adaptation of an ancestral actinobacterium to thermophily, as discussed in detail by Cavalier- Smith (2002a). 4 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. II phase: In the next phase, archaebacteria and eukaryotes diverged dramatically. Archaebacteria retained the wall and therefore their general bacterial cell and genetic organization, but became adapted to even hotter and more acidic environments by substituting prenyl ether lipids for the ancestral acyl esters and making new acid- resistant flagellar shafts (Cavalier-Smith, 2002a). -

Protist Phylogeny and the High-Level Classification of Protozoa
Europ. J. Protistol. 39, 338–348 (2003) © Urban & Fischer Verlag http://www.urbanfischer.de/journals/ejp Protist phylogeny and the high-level classification of Protozoa Thomas Cavalier-Smith Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK; E-mail: [email protected] Received 1 September 2003; 29 September 2003. Accepted: 29 September 2003 Protist large-scale phylogeny is briefly reviewed and a revised higher classification of the kingdom Pro- tozoa into 11 phyla presented. Complementary gene fusions reveal a fundamental bifurcation among eu- karyotes between two major clades: the ancestrally uniciliate (often unicentriolar) unikonts and the an- cestrally biciliate bikonts, which undergo ciliary transformation by converting a younger anterior cilium into a dissimilar older posterior cilium. Unikonts comprise the ancestrally unikont protozoan phylum Amoebozoa and the opisthokonts (kingdom Animalia, phylum Choanozoa, their sisters or ancestors; and kingdom Fungi). They share a derived triple-gene fusion, absent from bikonts. Bikonts contrastingly share a derived gene fusion between dihydrofolate reductase and thymidylate synthase and include plants and all other protists, comprising the protozoan infrakingdoms Rhizaria [phyla Cercozoa and Re- taria (Radiozoa, Foraminifera)] and Excavata (phyla Loukozoa, Metamonada, Euglenozoa, Percolozoa), plus the kingdom Plantae [Viridaeplantae, Rhodophyta (sisters); Glaucophyta], the chromalveolate clade, and the protozoan phylum Apusozoa (Thecomonadea, Diphylleida). Chromalveolates comprise kingdom Chromista (Cryptista, Heterokonta, Haptophyta) and the protozoan infrakingdom Alveolata [phyla Cilio- phora and Miozoa (= Protalveolata, Dinozoa, Apicomplexa)], which diverged from a common ancestor that enslaved a red alga and evolved novel plastid protein-targeting machinery via the host rough ER and the enslaved algal plasma membrane (periplastid membrane). -

Using Diatom and Apicomplexan Models to Study the Heme Pathway of Chromera Velia
International Journal of Molecular Sciences Article Using Diatom and Apicomplexan Models to Study the Heme Pathway of Chromera velia Jitka Richtová 1,2, Lilach Sheiner 3 , Ansgar Gruber 1 , Shun-Min Yang 1,2 , LudˇekKoˇrený 4, Boris Striepen 5 and Miroslav Oborník 1,2,* 1 Biology Centre CAS, Laboratory of Evolutionary Protistology, Institute of Parasitology, 370 05 Ceskˇ é Budˇejovice,Czech Republic; [email protected] (J.R.); [email protected] (A.G.); [email protected] (S.-M.Y.) 2 Faculty of Science, University of South Bohemia, 370 05 Ceskˇ é Budˇejovice,Czech Republic 3 Welcome Centre for Integrative Parasitology, College of Medical, Veterinary and Life Sciences, Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8QQ, UK; [email protected] 4 Department of Biochemistry, University of Cambridge, Cambridge CB2 1TN, UK; [email protected] 5 Department of Pathobiology, School of Veterinary Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; [email protected] * Correspondence: [email protected]; Tel.: +420-387-775-464 Abstract: Heme biosynthesis is essential for almost all living organisms. Despite its conserved function, the pathway’s enzymes can be located in a remarkable diversity of cellular compartments in different organisms. This location does not always reflect their evolutionary origins, as might be expected from the history of their acquisition through endosymbiosis. Instead, the final subcellular localization of the enzyme reflects multiple factors, including evolutionary origin, demand for the product, availability of the substrate, and mechanism of pathway regulation. The biosynthesis of Citation: Richtová, J.; Sheiner, L.; heme in the apicomonad Chromera velia follows a chimeric pathway combining heme elements from Gruber, A.; Yang, S.-M.; Koˇrený,L.; the ancient algal symbiont and the host. -

Extensive Molecular Tinkering in the Evolution of the Membrane Attachment Mode of the Rheb Gtpase
www.nature.com/scientificreports OPEN Extensive molecular tinkering in the evolution of the membrane attachment mode of the Rheb Received: 14 December 2017 Accepted: 15 March 2018 GTPase Published: xx xx xxxx Kristína Záhonová1, Romana Petrželková1, Matus Valach 2, Euki Yazaki3, Denis V. Tikhonenkov4, Anzhelika Butenko1, Jan Janouškovec5, Štěpánka Hrdá6, Vladimír Klimeš1, Gertraud Burger 2, Yuji Inagaki7, Patrick J. Keeling8, Vladimír Hampl6, Pavel Flegontov1, Vyacheslav Yurchenko1 & Marek Eliáš1 Rheb is a conserved and widespread Ras-like GTPase involved in cell growth regulation mediated by the (m)TORC1 kinase complex and implicated in tumourigenesis in humans. Rheb function depends on its association with membranes via prenylated C-terminus, a mechanism shared with many other eukaryotic GTPases. Strikingly, our analysis of a phylogenetically rich sample of Rheb sequences revealed that in multiple lineages this canonical and ancestral membrane attachment mode has been variously altered. The modifcations include: (1) accretion to the N-terminus of two diferent phosphatidylinositol 3-phosphate-binding domains, PX in Cryptista (the fusion being the frst proposed synapomorphy of this clade), and FYVE in Euglenozoa and the related undescribed fagellate SRT308; (2) acquisition of lipidic modifcations of the N-terminal region, namely myristoylation and/ or S-palmitoylation in seven diferent protist lineages; (3) acquisition of S-palmitoylation in the hypervariable C-terminal region of Rheb in apusomonads, convergently to some other Ras family proteins; (4) replacement of the C-terminal prenylation motif with four transmembrane segments in a novel Rheb paralog in the SAR clade; (5) loss of an evident C-terminal membrane attachment mechanism in Tremellomycetes and some Rheb paralogs of Euglenozoa. -

The Fungi Belonging to Kingdom Chromista: A. Oomycota
The Fungi belonging to Kingdom Chromista: a. Oomycota: Dr. Basudha Sharma Chromista ► Wittaker divided living beings into 5-Kingdom-Monera (Prokaryotic), Protista (Eukaryotic), Fungi, Plantae and Animalia. Molecular phylogeny, however suggested a new division: Chromista ► The chromists represent an independent evolutionary lineage that appears to have diverged from the same common ancestor as plants, animals and fungi. ► Chromista means ‘coloured’ and includes some colourless chromists like oomycota and certain algae. ► they all possess flagellated cells at some stage of their life cycles, and the flagella are typically of two types-hetrokont (whiplash and tinsel) ► the chloroplast is bounded by a double membrane, but has an extra layer of ER (also two-layered) that is often continuous with the nuclear envelope Chromista Oomycota ► commonly known as water moulds ► some are unicellular, however majority are multicellular and mycelial (branched filamentous coenocyte) ► They are classified as chromists because their free-swimming zoospores possess the heterokont-type flagella also, reserve food is stored in the form of mycolaminarin, an energy storage molecule similar to that found in diatoms and brown algae. ► oomycetes cell walls are composed of cellulose, free-living stage of the oomycetes has a diploid chromosome complement while that of the fungi is haploid. ► Asexual reproduction is by biflagellate zoospores ► Sexual reproduction is oogamus and involves the formation of oogonia and antheridia Saprolegnia ► These fungi are saprophytic organisms that are widely distributed in the aquatic environment and can derive nutrients from any organic source in water ► They become pathogenic to fish only when fish are stressed or eg. diseased. They attach to surfaces like gills and fins of fishes ► Reproduction in Saprolegnia may be sexual or asexual. -

Diptera: Sciaroidea Incertae Sedis), with Description of Three New Species from India and Vietnam
ACTA ENTOMOLOGICA MUSEI NATIONALIS PRAGAE Published 15.xii.2014 Volume 54(2), pp. 729–739 ISSN 0374-1036 http://zoobank.org/urn:lsid:zoobank.org:pub:8FF046B2-6267-454D-8A28-9AE80CC0F23C Notes on Nepaletricha (Diptera: Sciaroidea incertae sedis), with description of three new species from India and Vietnam Heikki HIPPA1) & Jan ŠEVýÍK2) 1) Gribbylunds allé 2, SE-183 65 Täby, Sweden; e-mail: [email protected] 2) Department of Biology and Ecology, University of Ostrava, Chittussiho 10, CZ-71000 Ostrava, Czech Republic; e-mail: [email protected] Abstract. The following new species are described: Nepaletricha dembickyi sp. nov. (India), N. lobosa sp. nov. (Vietnam) and N. sigma sp. nov. (India). The sys- tematic position of the genus is brieÀ y discussed. A key to the six known species of Nepaletricha is given. Key words. Insecta, Diptera, Sciaroidea, Antefungivoridae, Pleciomimidae, Rangomaramidae, Heterotricha group, Nepaletricha, fungus gnats, taxonomy, key, Oriental Region Introduction The genus Nepaletricha was established by CHANDLER (2002) for a single Oriental species Nepaletricha mystica Chandler, 2002. Subsequently, HIPPA et al. (2009) re-characterized the genus and added two new species from northern Thailand and Vietnam, respectively. The genus was considered as Sciaroidea incertae sedis by CHANDLER (2002) and HIPPA & VILKAMAA (2005, 2006). AMORIM & RINDAL (2007) placed Nepaletricha in the family Rangomaramidae, together with many other earlier unplaced genera of Sciaroidea. Concerning Nepaletricha, HIPPA et al. (2009) followed this placement. The classi¿ cation of AMORIM & RINDAL (2007) was strongly criticized by JASCHHOF (2011) who did not accept the new concept of Rangomara- midae and suggested to continue using Sciaroidea incertae sedis for a number of problematic genera of Sciaroidea. -

Conicocassis, a New Genus of Arcellinina (Testate Lobose Amoebae)
Palaeontologia Electronica palaeo-electronica.org Conicocassis, a new genus of Arcellinina (testate lobose amoebae) Nawaf A. Nasser and R. Timothy Patterson ABSTRACT Superfamily Arcellinina (informally known as thecamoebians or testate lobose amoebae) are a group of shelled benthic protists common in most Quaternary lacus- trine sediments. They are found worldwide, from the equator to the poles, living in a variety of fresh to brackish aquatic and terrestrial habitats. More than 130 arcellininid species and strains are ascribed to the genus Centropyxis Stein, 1857 within the family Centropyxidae Jung, 1942, which includes species that are distinguished by having a dorsoventral-oriented and flattened beret-like test (shell). Conicocassis, a new arcel- lininid genus of Centropyxidae differs from other genera of the family, specifically genus Centropyxis and its type species C. aculeata (Ehrenberg, 1932), by having a unique test comprised of two distinct components; a generally ovoid to subspherical, dorsoventral-oriented test body, with a pronounced asymmetrically positioned, funnel- like flange extending from a small circular aperture. The type species of the new genus, Conicocassis pontigulasiformis (Beyens et al., 1986) has previously been reported from peatlands in Germany, the Netherlands and Austria, as well as very wet mosses and aquatic environments in High Arctic regions of Europe and North America. The occurrence of the species in lacustrine environments in the central Northwest Ter- ritories extends the known geographic distribution of the genus in North America con- siderably southward. Nawaf A. Nasser. Department of Earth Sciences, Carleton University, 1125 Colonel By Drive, Ottawa, Ontario, K1S 5B6, Canada. [email protected] R. Timothy Patterson.