The Soil Microbial Food Web Revisited: Predatory Myxobacteria As Keystone Taxa?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Two Independent Retrons with Highly Diverse Reverse Transcriptases In
Proc. Natl. Acad. Sci. USA Vol. 87, pp. 942-945, February 1990 Biochemistry Two independent retrons with highly diverse reverse transcriptases in Myxococcus xanthus (multicopy single-stranded DNA/2',5'-phosphodiester/codon usage/myxobacteria) SUMIKO INOUYE, PETER J. HERZER, AND MASAYORI INOUYE Department of Biochemistry, University of Medicine and Dentistry of New Jersey, Robert Wood Johnson Medical School, Piscataway, NJ 08854 Communicated by Russell F. Doolittle, November 27, 1989 (received for review September 29, 1989) ABSTRACT A reverse transcriptase (RT) was recently found that the Mx65 retron is highly diverse and independent found in Myxococcus xanthus, a Gram-negative soil bacterium. from the Mx162 retron and that RT associated with the Mx65 This RT has been shown to be associated with a chromosomal retron has only 47% identity with RT for the Mx162 retron. region designated a retron responsible for the synthesis of a peculiar extrachromosomal DNA called msDNA (multicopy single-stranded DNA). We demonstrate that M. xanthus con- MATERIALS AND METHODS tains two independent, unlinked retrons, one for the synthesis Materials. The clone of the 9.0-kilobase (kb) Pst I fragment of msDNA-Mxl62 and the other for msDNA-Mx65. The struc- containing the Mx65 retron was obtained (8). Restriction tural analysis of the retron for msDNA-Mx65 revealed that the enzymes were purchased from New England Biolabs and coding regions for msdRNA (msr) and msDNA (msd), and an Boehringer Mannheim. open reading frame (ORF) downstream of msr are arranged in A deletion mutant strain ofthe Mx162 retron, AmsSX, was the same manner as found for the Mx162 retron. -

CUED Phd and Mphil Thesis Classes
High-throughput Experimental and Computational Studies of Bacterial Evolution Lars Barquist Queens' College University of Cambridge A thesis submitted for the degree of Doctor of Philosophy 23 August 2013 Arrakis teaches the attitude of the knife { chopping off what's incomplete and saying: \Now it's complete because it's ended here." Collected Sayings of Muad'dib Declaration High-throughput Experimental and Computational Studies of Bacterial Evolution The work presented in this dissertation was carried out at the Wellcome Trust Sanger Institute between October 2009 and August 2013. This dissertation is the result of my own work and includes nothing which is the outcome of work done in collaboration except where specifically indicated in the text. This dissertation does not exceed the limit of 60,000 words as specified by the Faculty of Biology Degree Committee. This dissertation has been typeset in 12pt Computer Modern font using LATEX according to the specifications set by the Board of Graduate Studies and the Faculty of Biology Degree Committee. No part of this dissertation or anything substantially similar has been or is being submitted for any other qualification at any other university. Acknowledgements I have been tremendously fortunate to spend the past four years on the Wellcome Trust Genome Campus at the Sanger Institute and the European Bioinformatics Institute. I would like to thank foremost my main collaborators on the studies described in this thesis: Paul Gardner and Gemma Langridge. Their contributions and support have been invaluable. I would also like to thank my supervisor, Alex Bateman, for giving me the freedom to pursue a wide range of projects during my time in his group and for advice. -

First Genomic Insights Into Members of a Candidate Bacterial Phylum Responsible for Wastewater Bulking
First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking Yuji Sekiguchi1, Akiko Ohashi1, Donovan H. Parks2, Toshihiro Yamauchi3, Gene W. Tyson2,4 and Philip Hugenholtz2,5 1 Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Ibaraki, Japan 2 Australian Centre for Ecogenomics, School of Chemistry and Molecular Biosciences, The University of Queensland, St. Lucia, Queensland, Australia 3 Administrative Management Department, Kubota Kasui Corporation, Minato-ku, Tokyo, Japan 4 Advanced Water Management Centre, The University of Queensland, St. Lucia, Queensland, Australia 5 Institute for Molecular Bioscience, The University of Queensland, St. Lucia, Queensland, Australia ABSTRACT Filamentous cells belonging to the candidate bacterial phylum KSB3 were previously identified as the causative agent of fatal filament overgrowth (bulking) in a high-rate industrial anaerobic wastewater treatment bioreactor. Here, we obtained near complete genomes from two KSB3 populations in the bioreactor, including the dominant bulking filament, using diVerential coverage binning of metagenomic data. Fluorescence in situ hybridization with 16S rRNA-targeted probes specific for the two populations confirmed that both are filamentous organisms. Genome-based metabolic reconstruction and microscopic observation of the KSB3 filaments in the presence of sugar gradients indicate that both filament types are Gram-negative, strictly anaerobic fermenters capable of -

Scope of the Thesis
Unraveling predatory-prey interactions between bacteria Dissertation To Fulfill the Requirements for the Degree of „Doctor rerum naturalium“ (Dr. rer. nat.) Submitted to the Council of the Faculty of Biology and Pharmacy of the Friedrich Schiller University Jena By Ivana Seccareccia (M.Sc. in Biology) born on 8th April 1986 in Rijeka, Croatia Die Forschungsarbeit im Rahmen dieser Dissertation wurde am Leibniz-Institut für Naturstoff-Forschung und Infektionsbiologie e.V. – Hans-Knöll–Institut in der Nachwuchsgruppe Sekundärmetabolismus räuberischer Bakterien unter der Betreuung von Dr. habil. Markus Nett von Oktober 2011 bis Oktober 2015 in Jena durchgeführt. Gutachter: ……………………………………………. ………………………………………….… ……………………………………………. Tag der öffentlichen Verteidigung: We make our world significant by the courage of our questions and by the depth of our answers. Carl Sagan Table of Contents 1 Introduction ......................................................................................................................... 6 1.1 Predation in the microbial community ........................................................................... 6 1.2 Bacterial predators .......................................................................................................... 7 1.3 Phases of predation ......................................................................................................... 8 1.3.1 Seeking prey ......................................................................................................... 9 1.3.2 Prey recognition -

The Phagotrophic Origin of Eukaryotes and Phylogenetic Classification Of
International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 DOI: 10.1099/ijs.0.02058-0 The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa Department of Zoology, T. Cavalier-Smith University of Oxford, South Parks Road, Oxford OX1 3PS, UK Tel: j44 1865 281065. Fax: j44 1865 281310. e-mail: tom.cavalier-smith!zoo.ox.ac.uk Eukaryotes and archaebacteria form the clade neomura and are sisters, as shown decisively by genes fragmented only in archaebacteria and by many sequence trees. This sisterhood refutes all theories that eukaryotes originated by merging an archaebacterium and an α-proteobacterium, which also fail to account for numerous features shared specifically by eukaryotes and actinobacteria. I revise the phagotrophy theory of eukaryote origins by arguing that the essentially autogenous origins of most eukaryotic cell properties (phagotrophy, endomembrane system including peroxisomes, cytoskeleton, nucleus, mitosis and sex) partially overlapped and were synergistic with the symbiogenetic origin of mitochondria from an α-proteobacterium. These radical innovations occurred in a derivative of the neomuran common ancestor, which itself had evolved immediately prior to the divergence of eukaryotes and archaebacteria by drastic alterations to its eubacterial ancestor, an actinobacterial posibacterium able to make sterols, by replacing murein peptidoglycan by N-linked glycoproteins and a multitude of other shared neomuran novelties. The conversion of the rigid neomuran wall into a flexible surface coat and the associated origin of phagotrophy were instrumental in the evolution of the endomembrane system, cytoskeleton, nuclear organization and division and sexual life-cycles. Cilia evolved not by symbiogenesis but by autogenous specialization of the cytoskeleton. -

Isolation, Antimicrobial Activity of Myxobacterial Crude Extracts and Identification of the Most Potent Strains
Arch Biol Sci. 2017;69(3):561-568 https://doi.org/10.2298/ABS161011132C Isolation, antimicrobial activity of myxobacterial crude extracts and identification of the most potent strains Ivana Charousová*, Juraj Medo and Soňa Javoreková Department of Microbiology, Faculty of Biotechnology and Food Sciences, Slovak University of Agriculture in Nitra, Tr. A. Hlinku 2, 949 76 Nitra, Slovakia *Corresponding author: [email protected] Received: October 11, 2016; Revised: November 14, 2016; Accepted: November 18, 2016; Published online: December 14, 2016 Abstract: Broad spectrum antimicrobial agents are urgently needed to fight frequently occurring multidrug-resistant pathogens. Myxobacteria have been regarded as “microbe factories” for active secondary metabolites, and therefore, this study was performed to isolate two bacteriolytic genera of myxobacteria, Myxococcus sp. and Corallococcus sp., from 10 soil/sand samples using two conventional methods followed by purification with the aim of determining the antimicrobial activity of methanol extracts against 11 test microorganisms (four Gram-positive, four Gram-negative, two yeasts and one fungus). Out of thirty-nine directly observed strains, 23 were purified and analyzed for antimicrobial activities. Based on the broth microdilution method, a total of 19 crude extracts showed antimicrobial activity. The range of inhibited wells was more important in the case of anti-Gram-positive-bacterial activity in comparison with the anti-Gram-negative-bacterial and antifungal activity. In light of the established degree and range of antimicrobial activity, two of the most active isolates (BNEM1 and SFEC2) were selected for further characterization. Morphological parameters and a sequence similarity search by BLAST revealed that they showed 99% sequence similarity to Myxococcus xanthus − BNEM1 (accession no. -
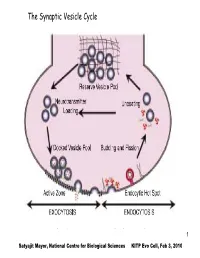
The Synaptic Vesicle Cycle
The Synaptic Vesicle Cycle 1 Satyajit Mayor, National Centre for Biological Sciences KITP Evo Cell, Feb 3, 2010 The Eukaryotic Commandments: T. Cavalier-Smith (i)origin of the endomembrane system (ER, Golgi and lysosomes) and coated- vesicle budding and fusion, including endocytosis and exocytosis; (ii) origin of the cytoskeleton, centrioles, cilia and associated molecular motors; (iii) origin of the nucleus, nuclear pore complex and trans-envelope protein and RNA transport; (iv) origin of linear chromosomes with plural replicons, centromeres and telomeres; (v) origin of novel cell-cycle controls and mitotic segregation; (vi) origin of sex (syngamy, nuclear fusion and meiosis); (vii) origin of peroxisomes; (ix) origin of mitochondria; (viii) novel patterns of rRNAprocessing using small nucleolar-ribonucleoproteins (snoRNPs); (x) origin of spliceosomal introns. 2 EM of Chemical synapse mitochondria Active Zone Figure 5.3, Bear, 2001 3 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. I phase: The ancestors of eukaryotes, the stem neomura, are shared with archaebacteria and evolved during the neomuran revolution, in which N-linked glycoproteins replaced murein peptidoglycan and 18 other suites of characters changed radically through adaptation of an ancestral actinobacterium to thermophily, as discussed in detail by Cavalier- Smith (2002a). 4 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. II phase: In the next phase, archaebacteria and eukaryotes diverged dramatically. Archaebacteria retained the wall and therefore their general bacterial cell and genetic organization, but became adapted to even hotter and more acidic environments by substituting prenyl ether lipids for the ancestral acyl esters and making new acid- resistant flagellar shafts (Cavalier-Smith, 2002a). -

Lysobacter Enzymogenes
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Dissertations and Theses in Biological Sciences Biological Sciences, School of 8-2010 Detection Methods for the Genus Lysobacter and the Species Lysobacter enzymogenes Hu Yin University of Nebraska at Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/bioscidiss Part of the Life Sciences Commons Yin, Hu, "Detection Methods for the Genus Lysobacter and the Species Lysobacter enzymogenes" (2010). Dissertations and Theses in Biological Sciences. 15. https://digitalcommons.unl.edu/bioscidiss/15 This Article is brought to you for free and open access by the Biological Sciences, School of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Dissertations and Theses in Biological Sciences by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Detection Methods for the Genus Lysobacter and the Species Lysobacter enzymogenes By Hu Yin A THESIS Presented to the Faculty of The Graduate College at the University of Nebraska In Partial Fulfillment of Requirements For the Degree of Master of Science Major: Biological Sciences Under the Supervision of Professor Gary Y. Yuen Lincoln, Nebraska August, 2010 Detection Methods for the Genus Lysobacter and the Species Lysobacter enzymogenes Hu Yin, M.S. University of Nebraska, 2010 Advisor: Gary Y. Yuen Strains of Lysobacter enzymogenes, a bacterial species with biocontrol activity, have been detected via 16S rDNA sequences in soil in different parts of the world. In most instances, however, their occurrence could not be confirmed by isolation, presumably because the species occurred in low numbers relative to faster-growing species of Bacillus or Pseudomonas. -

Thèse Morgane Wartel
Faculté des Sciences - Aix-Marseille Université Laboratoire de Chimie Bactérienne 163 Avenue de Luminy - Case 901 31 Chemin Joseph Aiguier 13288 Marseille 13009 Marseille Thèse En vue d’obtenir le grade de Docteur d’Aix-Marseille Université Biologie, spécialité Microbiologie Présentée et soutenue publiquement par Morgane Wartel Le Mercredi 18 Décembre 2013 A novel class of bacterial motors involved in the directional transport of a sugar at the bacterial surface: The machineries of motility and sporulation in Myxococcus xanthus. Une nouvelle classe de moteurs bactériens impliqués dans le transport de macromolécules à la surface bactérienne: Les machineries de motilité et de sporulation de Myxococcus xanthus. Membres du Jury : Dr. Christophe Grangeasse Rapporteur Dr. Patrick Viollier Rapporteur Pr. Pascale Cossart Examinateur Dr. Francis-André Wollman Examinateur Dr. Tâm Mignot Directeur de Thèse Pr. Frédéric Barras Président du Jury Faculté des Sciences - Aix-Marseille Université Laboratoire de Chimie Bactérienne 163 Avenue de Luminy - Case 901 31 Chemin Joseph Aiguier 13288 Marseille 13009 Marseille Thèse En vue d’obtenir le grade de Docteur d’Aix-Marseille Université Biologie, spécialité Microbiologie Présentée et soutenue publiquement par Morgane Wartel Le Mercredi 18 Décembre 2013 A novel class of bacterial motors involved in the directional transport of a sugar at the bacterial surface: The machineries of motility and sporulation in Myxococcus xanthus. Une nouvelle classe de moteurs bactériens impliqués dans le transport de macromolécules à la surface bactérienne: Les machineries de motilité et de sporulation de Myxococcus xanthus. Membres du Jury : Dr. Christophe Grangeasse Rapporteur Dr. Patrick Viollier Rapporteur Pr. Pascale Cossart Examinateur Dr. Francis-André Wollman Examinateur Dr. -

The Myxocoumarins a and B from Stigmatella Aurantiaca Strain MYX-030
The myxocoumarins A and B from Stigmatella aurantiaca strain MYX-030 Tobias A. M. Gulder*1, Snežana Neff2, Traugott Schüz2, Tammo Winkler2, René Gees2 and Bettina Böhlendorf*2 Full Research Paper Open Access Address: Beilstein J. Org. Chem. 2013, 9, 2579–2585. 1Kekulé Institute of Organic Chemistry and Biochemistry, University of doi:10.3762/bjoc.9.293 Bonn, Gerhard-Domagk-Straβe 1, 53121 Bonn, Germany and 2Syngenta Crop Protection AG, CH-4002 Basel, Switzerland Received: 21 August 2013 Accepted: 01 November 2013 Email: Published: 20 November 2013 Tobias A. M. Gulder* - [email protected]; Bettina Böhlendorf* - [email protected] This article is part of the Thematic Series "Natural products in synthesis and biosynthesis" and is dedicated to Prof. Dr. Gerhard Höfle on the * Corresponding author occasion of his 74th birthday. Keywords: Guest Editor: J. S. Dickschat antifungal activity; myxobacteria; natural products; Stigmatella aurantiaca; structure elucidation © 2013 Gulder et al; licensee Beilstein-Institut. License and terms: see end of document. Abstract The myxobacterial strain Stigmatella aurantiaca MYX-030 was selected as promising source for the discovery of new biologically active natural products by our screening methodology. The isolation, structure elucidation and initial biological evaluation of the myxocoumarins derived from this strain are described in this work. These compounds comprise an unusual structural framework and exhibit remarkable antifungal properties. Introduction Despite declining interest of most big R&D-driven chemical epothilones A (1), Figure 1), microtubule-stabilizing macrolac- companies in recent years, natural products continue to serve as tones that are clinically used in cancer therapy [16-20]. one of the most important sources of new bioactive chemical entities, both in the pharmaceutical [1-4] as well as in the agro- But also from an agrochemical point of view, myxobacteria chemical industry [5-8]. -

Emended Descriptions of the Genera Myxococcus and Corallococcus, Typification of the Species Myxococcus Stipitatus and Myxococcu
International Journal of Systematic and Evolutionary Microbiology (2009), 59, 2122–2128 DOI 10.1099/ijs.0.003566-0 Emended descriptions of the genera Myxococcus and Corallococcus, typification of the species Myxococcus stipitatus and Myxococcus macrosporus and a proposal that they be represented by neotype strains. Request for an Opinion Elke Lang and Erko Stackebrandt Correspondence DSMZ – Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Inhoffenstr. Elke Lang 7B, 38124 Braunschweig Germany [email protected] The genus Corallococcus was separated from the genus Myxococcus mainly on the basis of differences in morphology and consistency of swarms and fruiting bodies of the respective members. Phylogenetic, chemotaxonomic and physiological evidence is presented here that underpins the separate status of these phylogenetically neighbouring genera. Emended descriptions of the two genera are presented. The data also suggest that the species Corallococcus macrosporus belongs to the genus Myxococcus. To the best of our knowledge, the type strains of the species Myxococcus macrosporus and Myxococcus stipitatus are not available from any established culture collection or any other source. A Request for an Opinion is made regarding the proposal that strain Cc m8 (5DSM 14697 5CIP 109128) be formally recognized as the neotype strain for the species Myxococcus macrosporus, replacing the designated type strain Windsor M271T, and that strain Mx s8 (5DSM 14675 5JCM 12634) be formally recognized as the neotype strain for the species Myxococcus stipitatus, replacing the designated type strain Windsor M78T. Introduction The genus Myxococcus was described by Thaxter (1892), Within the family Myxococcaceae, delineation of genera, who first recognized that the myxobacteria, formerly descriptions of species and affiliations of strains to species assigned to the fungi, were in fact bacteria.