Taxonomy (Biology)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

History and Philosophy of Systematic Biology
History and Philosophy of Systematic Biology Bock, W. J. (1973) Philosophical foundations of classical evolutionary classification Systematic Zoology 22: 375-392 Part of a general symposium on "Contemporary Systematic Philosophies," there are some other interesting papers here. Brower, A. V. Z. (2000) Evolution Is Not a Necessary Assumption of Cladistics Cladistics 16: 143- 154 Dayrat, Benoit (2005) Ancestor-descendant relationships and the reconstruction of the Tree of Lif Paleobiology 31: 347-353 Donoghue, M.J. and J.W. Kadereit (1992) Walter Zimmermann and the growth of phylogenetic theory Systematic Biology 41: 74-84 Faith, D. P. and J. W. H. Trueman (2001) Towards an inclusive philosophy for phylogenetic inference Systematic Biology 50: 331-350 Gaffney, E. S. (1979) An introduction to the logic of phylogeny reconstruction, pp. 79-111 in Cracraft, J. and N. Eldredge (eds.) Phylogenetic Analysis and Paleontology Columbia University Press, New York. Gilmour, J. S. L. (1940) Taxonomy and philosophy, pp. 461-474 in J. Huxley (ed.) The New Systematics Oxford Hull, D. L. (1978) A matter of individuality Phil. of Science 45: 335-360 Hull, D. L. (1978) The principles of biological classification: the use and abuse of philosophy Hull, D. L. (1984) Cladistic theory: hypotheses that blur and grow, pp. 5-23 in T. Duncan and T. F. Stuessy (eds.) Cladistics: Perspectives on the Reconstruction of Evolutionary History Columbia University Press, New York * Hull, D. L. (1988) Science as a process: an evolutionary account of the social and conceptual development of science University of Chicago Press. An already classic work on the recent, violent history of systematics; used as data for Hull's general theories about scientific change. -

Mnemonic Memory Taxonomy
MNEMONIC MEMORY TAXONOMY Overview: In this lesson, students determine proper classification of organisms according to taxonomic levels, explore characteristics that determine classification, and create methods to recall ordered taxonomic terminology. Objectives: The student will: • describe the use and function of a taxonomy, specifically to order and classify living organisms; and • identify and list taxonomic levels of biological classification. Targeted Alaska Grade Level Expectations: Science [7] SC2.2 The student demonstrates an understanding of the structure, function, behavior, development, life cycles, and diversity of living organisms by identifying the seven levels of classification of organisms. [7] SA1.1 The student demonstrates an understanding of the processes of science by asking questions, predicting, observing, describing, measuring, classifying, makign generalizations, inferring, and communicating. Vocabulary: animalia— one of six kingdoms, including most living things that are able to move and digest food internally plantae –one of six kingdoms, including living things that generally manufacture their own food through the use of photosynthesis fungi – one of six kingdoms, mostly living things that are nonmobile and assist in the decomposition process, including yeasts, molds, and mushrooms binomial nomenclature – a scientific naming system that gives a unique name (“scientific name”) to each species, using organisms’ genus and species as its two parts (e.g., “Homo sapiens” for humans) class – a taxonomic rank below -

Supplementary Data Sterner, Beckett* and Scott Lidgard. (Under Revision
Supplementary Data Sterner, Beckett* and Scott Lidgard. (Under revision) “Moving Past the Systematics Wars.” Journal of the History of Biology. *Corresponding author E-mail: [email protected] Table S1 Examples of additive coding procedures used in conjuction with both phenetic & cladistic methodologies. Methods Taxa & Characters Use of additive Publication coding phenetic hyphomycetes fungi; phenetic Kendrick & Proctor morphology 1964 phenetic helminthosporium phenetic Ibrahim & Threlfall fungi; morphology, 1966 pathogenicity, physiology, biochemistry phenetic Listeria, Streptococci, phenetic Davis et al. 1969 related bacteria; morphology, biochemistry, sediment phenetic Agrobacterium; phenetic Kesters et al. 1973 morphology, biochemistry phenetic fossil nummulitid phenetic Barnett 1974 foraminifera; morphology phenetic anaerobic bacteria; phenetic Holmberg & Nord morphology, 1975 biochemistry phenetic actinomycete bacteria; phenetic Goodfellow et al. morphology, 1979 biochemistry phenetic freshwater sediment phenetic Mallory & Sayler bacteria; morphology, 1984 biochemistry phenetic nemerteans; phenetic Sundberg 1985 morphology, ecology, development phenetic rhodococci bacteria; phenetic Goodfellow et al. morphology, 1990 biochemistry phenetic actinomycete bacteria; phenetic O'Donnell et al. 1993 morphology, biochemistry phenetic Bacillus bacteria; phenetic Nielsen et al. 1995 biochemistry, DNA base composition phenetic fossil crinoids; skeletal phenetic Deline & Ausich 2011 morphology phenetic fossil crinoids; skeletal phenetic -

Antony Van Leeuwenhoek, the Father of Microscope
Turkish Journal of Biochemistry – Türk Biyokimya Dergisi 2016; 41(1): 58–62 Education Sector Letter to the Editor – 93585 Emine Elif Vatanoğlu-Lutz*, Ahmet Doğan Ataman Medicine in philately: Antony Van Leeuwenhoek, the father of microscope Pullardaki tıp: Antony Van Leeuwenhoek, mikroskobun kaşifi DOI 10.1515/tjb-2016-0010 only one lens to look at blood, insects and many other Received September 16, 2015; accepted December 1, 2015 objects. He was first to describe cells and bacteria, seen through his very small microscopes with, for his time, The origin of the word microscope comes from two Greek extremely good lenses (Figure 1) [3]. words, “uikpos,” small and “okottew,” view. It has been After van Leeuwenhoek’s contribution,there were big known for over 2000 years that glass bends light. In the steps in the world of microscopes. Several technical inno- 2nd century BC, Claudius Ptolemy described a stick appear- vations made microscopes better and easier to handle, ing to bend in a pool of water, and accurately recorded the which led to microscopy becoming more and more popular angles to within half a degree. He then very accurately among scientists. An important discovery was that lenses calculated the refraction constant of water. During the combining two types of glass could reduce the chromatic 1st century,around year 100, glass had been invented and effect, with its disturbing halos resulting from differences the Romans were looking through the glass and testing in refraction of light (Figure 2) [4]. it. They experimented with different shapes of clear glass In 1830, Joseph Jackson Lister reduced the problem and one of their samples was thick in the middle and thin with spherical aberration by showing that several weak on the edges [1]. -

Species Concepts and the Evolutionary Paradigm in Modern Nematology
JOURNAL OF NEMATOLOGY VOLUME 30 MARCH 1998 NUMBER 1 Journal of Nematology 30 (1) :1-21. 1998. © The Society of Nematologists 1998. Species Concepts and the Evolutionary Paradigm in Modern Nematology BYRON J. ADAMS 1 Abstract: Given the task of recovering and representing evolutionary history, nematode taxonomists can choose from among several species concepts. All species concepts have theoretical and (or) opera- tional inconsistencies that can result in failure to accurately recover and represent species. This failure not only obfuscates nematode taxonomy but hinders other research programs in hematology that are dependent upon a phylogenetically correct taxonomy, such as biodiversity, biogeography, cospeciation, coevolution, and adaptation. Three types of systematic errors inherent in different species concepts and their potential effects on these research programs are presented. These errors include overestimating and underestimating the number of species (type I and II error, respectively) and misrepresenting their phylogenetic relationships (type III error). For research programs in hematology that utilize recovered evolutionary history, type II and III errors are the most serious. Linnean, biological, evolutionary, and phylogenefic species concepts are evaluated based on their sensitivity to systematic error. Linnean and biologica[ species concepts are more prone to serious systematic error than evolutionary or phylogenetic concepts. As an alternative to the current paradigm, an amalgamation of evolutionary and phylogenetic species concepts is advocated, along with a set of discovery operations designed to minimize the risk of making systematic errors. Examples of these operations are applied to species and isolates of Heterorhab- ditis. Key words: adaptation, biodiversity, biogeography, coevolufion, comparative method, cospeciation, evolution, nematode, philosophy, species concepts, systematics, taxonomy. -

Western Tiger Salamander,Ambystoma Mavortium
COSEWIC Assessment and Status Report on the Western Tiger Salamander Ambystoma mavortium Southern Mountain population Prairie / Boreal population in Canada Southern Mountain population – ENDANGERED Prairie / Boreal population – SPECIAL CONCERN 2012 COSEWIC status reports are working documents used in assigning the status of wildlife species suspected of being at risk. This report may be cited as follows: COSEWIC. 2012. COSEWIC assessment and status report on the Western Tiger Salamander Ambystoma mavortium in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. xv + 63 pp. (www.registrelep-sararegistry.gc.ca/default_e.cfm). Previous report(s): COSEWIC. 2001. COSEWIC assessment and status report on the tiger salamander Ambystoma tigrinum in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. vi + 33 pp. (www.sararegistry.gc.ca/status/status_e.cfm). Schock, D.M. 2001. COSEWIC assessment and status report on the tiger salamander Ambystoma tigrinum in Canada, in COSEWIC assessment and status report on the tiger salamander Ambystoma tigrinum in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. 1-33 pp. Production note: COSEWIC would like to acknowledge Arthur Whiting for writing the status report on the Western Tiger Salamander, Ambystoma mavortium, in Canada, prepared under contract with Environment Canada. This report was overseen and edited by Kristiina Ovaska, Co-chair of the COSEWIC Amphibians and Reptiles Specialist Subcommittee. For additional copies contact: COSEWIC Secretariat c/o Canadian Wildlife Service Environment Canada Ottawa, ON K1A 0H3 Tel.: 819-953-3215 Fax: 819-994-3684 E-mail: COSEWIC/[email protected] http://www.cosewic.gc.ca Également disponible en français sous le titre Ếvaluation et Rapport de situation du COSEPAC sur la Salamandre tigrée de l’Ouest (Ambystoma mavortium) au Canada. -

Botanical Nomenclature: Concept, History of Botanical Nomenclature
Module – 15; Content writer: AvishekBhattacharjee Module 15: Botanical Nomenclature: Concept, history of botanical nomenclature (local and scientific) and its advantages, formation of code. Content writer: Dr.AvishekBhattacharjee, Central National Herbarium, Botanical Survey of India, P.O. – B. Garden, Howrah – 711 103. Module – 15; Content writer: AvishekBhattacharjee Botanical Nomenclature:Concept – A name is a handle by which a mental image is passed. Names are just labels we use to ensure we are understood when we communicate. Nomenclature is a mechanism for unambiguous communication about the elements of taxonomy. Botanical Nomenclature, i.e. naming of plants is that part of plant systematics dealing with application of scientific names to plants according to some set rules. It is related to, but distinct from taxonomy. A botanical name is a unique identifier to which information of a taxon can be attached, thus enabling the movement of data across languages, scientific disciplines, and electronic retrieval systems. A plant’s name permits ready summarization of information content of the taxon in a nested framework. A systemofnamingplantsforscientificcommunicationmustbe international inscope,andmustprovideconsistencyintheapplicationof names.Itmustalsobeacceptedbymost,ifnotall,membersofthe scientific community. These criteria led, almost inevitably, to International Botanical Congresses (IBCs) being the venue at which agreement on a system of scientific nomenclature for plants was sought. The IBCs led to publication of different ‘Codes’ which embodied the rules and regulations of botanical nomenclature and the decisions taken during these Congresses. Advantages ofBotanical Nomenclature: Though a common name may be much easier to remember, there are several good reasons to use botanical names for plant identification. Common names are not unique to a specific plant. -

Synthesis of Phylogeny and Taxonomy Into a Comprehensive Tree of Life
Synthesis of phylogeny and taxonomy into a comprehensive tree of life Cody E. Hinchliffa,1, Stephen A. Smitha,1,2, James F. Allmanb, J. Gordon Burleighc, Ruchi Chaudharyc, Lyndon M. Coghilld, Keith A. Crandalle, Jiabin Dengc, Bryan T. Drewf, Romina Gazisg, Karl Gudeh, David S. Hibbettg, Laura A. Katzi, H. Dail Laughinghouse IVi, Emily Jane McTavishj, Peter E. Midfordd, Christopher L. Owenc, Richard H. Reed, Jonathan A. Reesk, Douglas E. Soltisc,l, Tiffani Williamsm, and Karen A. Cranstonk,2 aEcology and Evolutionary Biology, University of Michigan, Ann Arbor, MI 48109; bInterrobang Corporation, Wake Forest, NC 27587; cDepartment of Biology, University of Florida, Gainesville, FL 32611; dField Museum of Natural History, Chicago, IL 60605; eComputational Biology Institute, George Washington University, Ashburn, VA 20147; fDepartment of Biology, University of Nebraska-Kearney, Kearney, NE 68849; gDepartment of Biology, Clark University, Worcester, MA 01610; hSchool of Journalism, Michigan State University, East Lansing, MI 48824; iBiological Science, Clark Science Center, Smith College, Northampton, MA 01063; jDepartment of Ecology and Evolutionary Biology, University of Kansas, Lawrence, KS 66045; kNational Evolutionary Synthesis Center, Duke University, Durham, NC 27705; lFlorida Museum of Natural History, University of Florida, Gainesville, FL 32611; and mComputer Science and Engineering, Texas A&M University, College Station, TX 77843 Edited by David M. Hillis, The University of Texas at Austin, Austin, TX, and approved July 28, 2015 (received for review December 3, 2014) Reconstructing the phylogenetic relationships that unite all line- published phylogenies are available only as journal figures, rather ages (the tree of life) is a grand challenge. The paucity of homologous than in electronic formats that can be integrated into databases and character data across disparately related lineages currently renders synthesis methods (7–9). -

The Phagotrophic Origin of Eukaryotes and Phylogenetic Classification Of
International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 DOI: 10.1099/ijs.0.02058-0 The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa Department of Zoology, T. Cavalier-Smith University of Oxford, South Parks Road, Oxford OX1 3PS, UK Tel: j44 1865 281065. Fax: j44 1865 281310. e-mail: tom.cavalier-smith!zoo.ox.ac.uk Eukaryotes and archaebacteria form the clade neomura and are sisters, as shown decisively by genes fragmented only in archaebacteria and by many sequence trees. This sisterhood refutes all theories that eukaryotes originated by merging an archaebacterium and an α-proteobacterium, which also fail to account for numerous features shared specifically by eukaryotes and actinobacteria. I revise the phagotrophy theory of eukaryote origins by arguing that the essentially autogenous origins of most eukaryotic cell properties (phagotrophy, endomembrane system including peroxisomes, cytoskeleton, nucleus, mitosis and sex) partially overlapped and were synergistic with the symbiogenetic origin of mitochondria from an α-proteobacterium. These radical innovations occurred in a derivative of the neomuran common ancestor, which itself had evolved immediately prior to the divergence of eukaryotes and archaebacteria by drastic alterations to its eubacterial ancestor, an actinobacterial posibacterium able to make sterols, by replacing murein peptidoglycan by N-linked glycoproteins and a multitude of other shared neomuran novelties. The conversion of the rigid neomuran wall into a flexible surface coat and the associated origin of phagotrophy were instrumental in the evolution of the endomembrane system, cytoskeleton, nuclear organization and division and sexual life-cycles. Cilia evolved not by symbiogenesis but by autogenous specialization of the cytoskeleton. -

Loggerhead Shrike, Migrans Subspecies (Lanius Ludovicianus Migrans), in Canada
PROPOSED Species at Risk Act Recovery Strategy Series Recovery Strategy for the Loggerhead Shrike, migrans subspecies (Lanius ludovicianus migrans), in Canada Loggerhead Shrike, migrans subspecies © Manitoba Conservation 2010 About the Species at Risk Act Recovery Strategy Series What is the Species at Risk Act (SARA)? SARA is the Act developed by the federal government as a key contribution to the common national effort to protect and conserve species at risk in Canada. SARA came into force in 2003, and one of its purposes is “to provide for the recovery of wildlife species that are extirpated, endangered or threatened as a result of human activity.” What is recovery? In the context of species at risk conservation, recovery is the process by which the decline of an endangered, threatened or extirpated species is arrested or reversed, and threats are removed or reduced to improve the likelihood of the species’ persistence in the wild. A species will be considered recovered when its long-term persistence in the wild has been secured. What is a recovery strategy? A recovery strategy is a planning document that identifies what needs to be done to arrest or reverse the decline of a species. It sets objectives and broad strategies to attain them and identifies the main areas of activities to be undertaken. Detailed planning is done at the action plan stage. Recovery strategy development is a commitment of all provinces and territories and of three federal agencies — Environment Canada, Parks Canada Agency and Fisheries and Oceans Canada — under the Accord for the Protection of Species at Risk. -

Biology Assessment Plan Spring 2019
Biological Sciences Department 1 Biology Assessment Plan Spring 2019 Task: Revise the Biology Program Assessment plans with the goal of developing a sustainable continuous improvement plan. In order to revise the program assessment plan, we have been asked by the university assessment committee to revise our Students Learning Outcomes (SLOs) and Program Learning Outcomes (PLOs). Proposed revisions Approach: A large community of biology educators have converged on a set of core biological concepts with five core concepts that all biology majors should master by graduation, namely 1) evolution; 2) structure and function; 3) information flow, exchange, and storage; 4) pathways and transformations of energy and matter; and (5) systems (Vision and Change, AAAS, 2011). Aligning our student learning and program goals with Vision and Change (V&C) provides many advantages. For example, the V&C community has recently published a programmatic assessment to measure student understanding of vision and change core concepts across general biology programs (Couch et al. 2019). They have also carefully outlined student learning conceptual elements (see Appendix A). Using the proposed assessment will allow us to compare our student learning profiles to those of similar institutions across the country. Revised Student Learning Objectives SLO 1. Students will demonstrate an understanding of core concepts spanning scales from molecules to ecosystems, by analyzing biological scenarios and data from scientific studies. Students will correctly identify and explain the core biological concepts involved relative to: biological evolution, structure and function, information flow, exchange, and storage, the pathways and transformations of energy and matter, and biological systems. More detailed statements of the conceptual elements students need to master are presented in appendix A. -
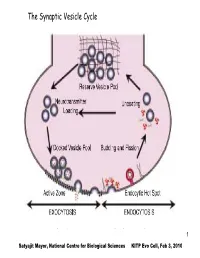
The Synaptic Vesicle Cycle
The Synaptic Vesicle Cycle 1 Satyajit Mayor, National Centre for Biological Sciences KITP Evo Cell, Feb 3, 2010 The Eukaryotic Commandments: T. Cavalier-Smith (i)origin of the endomembrane system (ER, Golgi and lysosomes) and coated- vesicle budding and fusion, including endocytosis and exocytosis; (ii) origin of the cytoskeleton, centrioles, cilia and associated molecular motors; (iii) origin of the nucleus, nuclear pore complex and trans-envelope protein and RNA transport; (iv) origin of linear chromosomes with plural replicons, centromeres and telomeres; (v) origin of novel cell-cycle controls and mitotic segregation; (vi) origin of sex (syngamy, nuclear fusion and meiosis); (vii) origin of peroxisomes; (ix) origin of mitochondria; (viii) novel patterns of rRNAprocessing using small nucleolar-ribonucleoproteins (snoRNPs); (x) origin of spliceosomal introns. 2 EM of Chemical synapse mitochondria Active Zone Figure 5.3, Bear, 2001 3 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. I phase: The ancestors of eukaryotes, the stem neomura, are shared with archaebacteria and evolved during the neomuran revolution, in which N-linked glycoproteins replaced murein peptidoglycan and 18 other suites of characters changed radically through adaptation of an ancestral actinobacterium to thermophily, as discussed in detail by Cavalier- Smith (2002a). 4 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. II phase: In the next phase, archaebacteria and eukaryotes diverged dramatically. Archaebacteria retained the wall and therefore their general bacterial cell and genetic organization, but became adapted to even hotter and more acidic environments by substituting prenyl ether lipids for the ancestral acyl esters and making new acid- resistant flagellar shafts (Cavalier-Smith, 2002a).