Comprehensive Review on the Clinical Relevance of Long Non-Coding Rnas in Cutaneous Melanoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Hidden Targets in RAF Signalling Pathways to Block Oncogenic RAS Signalling
G C A T T A C G G C A T genes Review Hidden Targets in RAF Signalling Pathways to Block Oncogenic RAS Signalling Aoife A. Nolan 1, Nourhan K. Aboud 1, Walter Kolch 1,2,* and David Matallanas 1,* 1 Systems Biology Ireland, School of Medicine, University College Dublin, Belfield, Dublin 4, Ireland; [email protected] (A.A.N.); [email protected] (N.K.A.) 2 Conway Institute of Biomolecular & Biomedical Research, University College Dublin, Belfield, Dublin 4, Ireland * Correspondence: [email protected] (W.K.); [email protected] (D.M.) Abstract: Oncogenic RAS (Rat sarcoma) mutations drive more than half of human cancers, and RAS inhibition is the holy grail of oncology. Thirty years of relentless efforts and harsh disappointments have taught us about the intricacies of oncogenic RAS signalling that allow us to now get a pharma- cological grip on this elusive protein. The inhibition of effector pathways, such as the RAF-MEK-ERK pathway, has largely proven disappointing. Thus far, most of these efforts were aimed at blocking the activation of ERK. Here, we discuss RAF-dependent pathways that are regulated through RAF functions independent of catalytic activity and their potential role as targets to block oncogenic RAS signalling. We focus on the now well documented roles of RAF kinase-independent functions in apoptosis, cell cycle progression and cell migration. Keywords: RAF kinase-independent; RAS; MST2; ASK; PLK; RHO-α; apoptosis; cell cycle; cancer therapy Citation: Nolan, A.A.; Aboud, N.K.; Kolch, W.; Matallanas, D. Hidden Targets in RAF Signalling Pathways to Block Oncogenic RAS Signalling. -

Table 2. Functional Classification of Genes Differentially Regulated After HOXB4 Inactivation in HSC/Hpcs
Table 2. Functional classification of genes differentially regulated after HOXB4 inactivation in HSC/HPCs Symbol Gene description Fold-change (mean ± SD) Signal transduction Adam8 A disintegrin and metalloprotease domain 8 1.91 ± 0.51 Arl4 ADP-ribosylation factor-like 4 - 1.80 ± 0.40 Dusp6 Dual specificity phosphatase 6 (Mkp3) - 2.30 ± 0.46 Ksr1 Kinase suppressor of ras 1 1.92 ± 0.42 Lyst Lysosomal trafficking regulator 1.89 ± 0.34 Mapk1ip1 Mitogen activated protein kinase 1 interacting protein 1 1.84 ± 0.22 Narf* Nuclear prelamin A recognition factor 2.12 ± 0.04 Plekha2 Pleckstrin homology domain-containing. family A. (phosphoinosite 2.15 ± 0.22 binding specific) member 2 Ptp4a2 Protein tyrosine phosphatase 4a2 - 2.04 ± 0.94 Rasa2* RAS p21 activator protein 2 - 2.80 ± 0.13 Rassf4 RAS association (RalGDS/AF-6) domain family 4 3.44 ± 2.56 Rgs18 Regulator of G-protein signaling - 1.93 ± 0.57 Rrad Ras-related associated with diabetes 1.81 ± 0.73 Sh3kbp1 SH3 domain kinase bindings protein 1 - 2.19 ± 0.53 Senp2 SUMO/sentrin specific protease 2 - 1.97 ± 0.49 Socs2 Suppressor of cytokine signaling 2 - 2.82 ± 0.85 Socs5 Suppressor of cytokine signaling 5 2.13 ± 0.08 Socs6 Suppressor of cytokine signaling 6 - 2.18 ± 0.38 Spry1 Sprouty 1 - 2.69 ± 0.19 Sos1 Son of sevenless homolog 1 (Drosophila) 2.16 ± 0.71 Ywhag 3-monooxygenase/tryptophan 5- monooxygenase activation protein. - 2.37 ± 1.42 gamma polypeptide Zfyve21 Zinc finger. FYVE domain containing 21 1.93 ± 0.57 Ligands and receptors Bambi BMP and activin membrane-bound inhibitor - 2.94 ± 0.62 -

TRANSCRIPTIONAL REGULATION of Hur in RENAL STRESS
TRANSCRIPTIONAL REGULATION OF HuR IN RENAL STRESS DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Sudha Suman Govindaraju Graduate Program in Biochemistry The Ohio State University 2014 Dissertation Committee: Dr. Beth S. Lee, Ph.D., Advisor Dr. Kathleen Boris-Lawrie, Ph.D. Dr. Sissy M. Jhiang, Ph.D. Dr. Arthur R. Strauch, Ph.D Abstract HuR is a ubiquitously expressed RNA-binding protein that affects the post- transcriptional life of thousands of cellular mRNAs by regulating transcript stability and translation. HuR can post-transcriptionally regulate gene expression and modulate cellular responses to stress, differentiation, proliferation, apoptosis, senescence, inflammation, and the immune response. It is an important mediator of survival during cellular stress, but when inappropriately expressed, can promote oncogenic transformation. Not surprisingly, the expression of HuR itself is tightly regulated at multiple transcriptional and post-transcriptional levels. Previous studies demonstrated the existence of two alternate HuR transcripts that differ in their 5’ untranslated regions and have markedly different translatabilities. These forms were also found to be reciprocally expressed following cellular stress in kidney proximal tubule cell lines, and the shorter, more readily translatable variant was shown to be regulated by Smad 1/5/8 pathway and bone morphogenetic protein-7 (BMP-7) signaling. In this study, the factors that promote transcription of the longer alternate form were identified. NF-κB was shown to be important for expression of the long HuR mRNA, as was a newly identified region with potential for binding the Sp/KLF families of transcription factors. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

CDK4/6 Inhibitors in Melanoma: a Comprehensive Review
cells Review CDK4/6 Inhibitors in Melanoma: A Comprehensive Review Mattia Garutti 1,*, Giada Targato 2 , Silvia Buriolla 2 , Lorenza Palmero 1,2 , Alessandro Marco Minisini 2 and Fabio Puglisi 1,2 1 CRO Aviano National Cancer Institute IRCCS, 33081 Aviano, Italy; [email protected] (L.P.); [email protected] (F.P.) 2 Department of Medicine (DAME), University of Udine, 33100 Udine, Italy; [email protected] (G.T.); [email protected] (S.B.); [email protected] (A.M.M.) * Correspondence: [email protected] Abstract: Historically, metastatic melanoma was considered a highly lethal disease. However, recent advances in drug development have allowed a significative improvement in prognosis. In particular, BRAF/MEK inhibitors and anti-PD1 antibodies have completely revolutionized the management of this disease. Nonetheless, not all patients derive a benefit or a durable benefit from these therapies. To overtake this challenges, new clinically active compounds are being tested in the context of clinical trials. CDK4/6 inhibitors are drugs already available in clinical practice and preliminary evidence showed a promising activity also in melanoma. Herein we review the available literature to depict a comprehensive landscape about CDK4/6 inhibitors in melanoma. We present the molecular and genetic background that might justify the usage of these drugs, the preclinical evidence, the clinical available data, and the most promising ongoing clinical trials. Keywords: CDK4/6; CDK4; CDK6; melanoma; Palbociclib; Ribociclib; Abemaciclib Citation: Garutti, M.; Targato, G.; Buriolla, S.; Palmero, L.; Minisini, A.M.; Puglisi, F. CDK4/6 Inhibitors in Melanoma: A Comprehensive 1. Introduction Review. Cells 2021, 10, 1334. -

Epigenetic Loss of the RNA Decapping Enzyme NUDT16 Mediates C-MYC Activation in T-Cell Acute Lymphoblastic Leukemia
OPEN Leukemia (2017) 31, 1622–1657 www.nature.com/leu LETTERS TO THE EDITOR Epigenetic loss of the RNA decapping enzyme NUDT16 mediates C-MYC activation in T-cell acute lymphoblastic leukemia Leukemia (2017) 31, 1622–1625; doi:10.1038/leu.2017.99 Having found the aforementioned NUDT16 CpG island methy- lation profiles, we studied in greater detail their association with the possible transcriptional inactivation of the NUDT16 gene at the RNA and protein levels in leukemia cell lines. We first It is possible that the occurrence of intrinsic defects in RNA performed bisulfite genomic sequencing of mutiple clones in the – processing pathways, such as RNA decapping,1 3 contribute to the T-cell Acute Lymphoblastic Leukemia (T-ALL) cell lines CCRF-CEM, distorted RNA landscapes of cancer cells. After transcription by Jurkat, MOLT-4 and MOLT-16 using primers that encompassed the RNA polymerase II, RNA molecules are equipped with a 5´-end transcription start site-associated CpG island and confirmed the N7-methyl guanosine (m7G)-cap. This m7G-cap is essential for hypermethylated status of the 5′-end region of NUDT16 in translation, stabilizing the RNA molecule and protecting it from comparison to normal T lymphocytes (Figure 1b), validating the – exonucleolytic breakdown.1 3 For RNA decay to occur the DNA methylation patterns obtained by the microarray approach m7G-cap first needs to be removed. This process is known as (Supplementary Figure S2). In contrast, normal T lymphocytes, the – decapping.1 3 The decapping mRNA 2 (DCP2) enzyme,4 also T-ALL cell lines KOPN-8, REH and RS4;11 and the leukemia cell known as the nucleoside diphosphate-linked moiety X motif lines HL-60 and K562 derived from myeloid lineage were all found 20 (NUDT20), was originally thought to be the only mammalian to be unmethylated (Figure 1b; Supplementary Figure S2). -
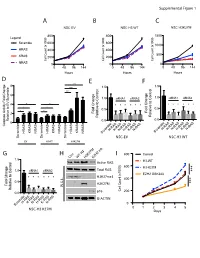
C a B D G E F
Supplemental Figure 1 A B C NSC EV NSC H3 WT NSC H3K27M 800 800 1500 ) ) Legend ) Scramble 600 600 x1000 x1000 x1000 ( ( ( 1000 t t t n n HRAS n 400 400 u u u o o o C C C 500 l l KRAS l 200 200 el el el C C C NRAS 0 0 0 0 48 96 144 0 48 96 144 0 48 96 144 Hours Hours Hours D *** E F 8 1.5 1.5 e *** l l *** ro ro t 6 t 1.0 siRNA1 siRNA2 1.0 siRNA1 siRNA2 Con Con o o t 4 * * t * * * * * * e * * * * * * e v i * * v i t 0.5 * * t 0.5 Fold Change 2 Fold Change Rela Rela Relative to EV Scrambl 0.0 0.0 0 Caspase Activity Fold Change H-RASK-RASN-RASH-RASK-RASN-RAS H-RASK-RASN-RASH-RASK-RASN-RAS KRAS KRAS KRAS HRAS NRAS HRAS NRAS HRAS NRAS Scramble Scramble Scramble Scramble Scramble NSC-EV NSC-H3 WT EV H3WT H3K27M G H I 800 Control Con WT H3 H3K27MEZH2 inb 1.5 l H3-WT Active RAS ro ) t 600 H3-K27M 1.0 siRNA1 siRNA2 Total RAS *** Con x1000 ( o EZH2 GSK343 t t * * * * * * H3K27me3 n e 400 u v i o t 0.5 *** H3K27M C Fold Change l W.C.L. el Rela p16 C 200 0.0 B-ACTIN H-RASK-RASN-RASH-RASK-RASN-RAS 0 Scramble NSC-H3 K27M 0 1 2 3 4 5 Days E C A 100 Cell Viability 100 25 50 75 Cell Viability 25 50 75 0 0 Fold Change MOCK MOCK Relative to Control Scramble * 0.0 0.2 0.4 0.6 0.8 1.0 1.2 MYC Scramble MYC MYC * PDGFRA Scramble siRNA PDGFRA MYC PDGFRA AURKA AURKA DIPG007siRNA2 PDGFRA NSC H3K27MsiRNA1and AURKA LAMTOR3 AURKA LAMTOR3 LAMTOR3 LIN28A LAMTOR3 LIN28A LIN28B LIN28A NSC-EV LIN28A MAP2K3 LIN28B LIN28B LIN28B MAP2K5 siRNA1 MAP2K3 MAP2K3 MAP3K2 MAP2K3 MAP3K7 MAP2K5 MAP2K5 MAPK7 MAP2K5 2 MAP3K2 siRNA1 siRNA2 MAP3K2 ZAK MAP3K7 MAP3K2 MAP3K7 MAPK7 MAP3K7 -

CDK4/6 Inhibition Reprograms Mitochondrial Metabolism in BRAFV600 Melanoma Via a P53 Dependent Pathway
cancers Article CDK4/6 Inhibition Reprograms Mitochondrial Metabolism in BRAFV600 Melanoma via a p53 Dependent Pathway Nancy T. Santiappillai 1,2, Shatha Abuhammad 1 , Alison Slater 1, Laura Kirby 1, Grant A. McArthur 1,3,†, Karen E. Sheppard 1,2,3,*,† and Lorey K. Smith 1,3,*,† 1 Cancer Research Division, Peter MacCallum Cancer Centre, Melbourne 3052, Australia; [email protected] (N.T.S.); [email protected] (S.A.); [email protected] (A.S.); [email protected] (L.K.); [email protected] (G.A.M.) 2 Department of Biochemistry and Molecular Biology, University of Melbourne, Melbourne 3052, Australia 3 Sir Peter MacCallum Department of Oncology, University of Melbourne, Melbourne 3052, Australia * Correspondence: [email protected] (K.E.S.); [email protected] (L.K.S.) † These authors contributed equally to the work. Simple Summary: Cyclin-dependent kinases 4 and 6 (CDK4/6) are key enzymes controlling the cell cycle. CDK4/6 inhibitors are being tested in multiple clinical trials for a range of cancers including melanoma, and a deeper understanding of how they interact with other therapies is vital for their clinical development. Beyond the cell cycle, CDK4/6 regulates cell metabolism, which is a critical factor determining response to standard-of-care mitogen-activated protein kinase (MAPK) pathway therapies in melanoma. Here, we show that CDK4/6 inhibitors increase glutamine and fatty acid-oxidation-dependent mitochondrial metabolism in melanoma cells, but they do not alter the metabolic response to MAPK inhibitors. These observations shed light on how CDK4/6 inhibitors Citation: Santiappillai, N.T.; impinge on the regulation of metabolism and how they interact with other therapies in the setting of Abuhammad, S.; Slater, A.; Kirby, L.; melanoma. -

A Dissertation Entitled the Androgen Receptor
A Dissertation entitled The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit Submitted to the Graduate Faculty as partial fulfillment of the requirements for the PhD Degree in Biomedical science Dr. Manohar Ratnam, Committee Chair Dr. Lirim Shemshedini, Committee Member Dr. Robert Trumbly, Committee Member Dr. Edwin Sanchez, Committee Member Dr. Beata Lecka -Czernik, Committee Member Dr. Patricia R. Komuniecki, Dean College of Graduate Studies The University of Toledo August 2011 Copyright 2011, Mesfin Gonit This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit As partial fulfillment of the requirements for the PhD Degree in Biomedical science The University of Toledo August 2011 Prostate cancer depends on the androgen receptor (AR) for growth and survival even in the absence of androgen. In the classical models of gene activation by AR, ligand activated AR signals through binding to the androgen response elements (AREs) in the target gene promoter/enhancer. In the present study the role of AREs in the androgen- independent transcriptional signaling was investigated using LP50 cells, derived from parental LNCaP cells through extended passage in vitro. LP50 cells reflected the signature gene overexpression profile of advanced clinical prostate tumors. The growth of LP50 cells was profoundly dependent on nuclear localized AR but was independent of androgen. Nevertheless, in these cells AR was unable to bind to AREs in the absence of androgen. -

In This Issue
IN THIS ISSUE AACR Project GENIE Facilitates Sharing of Genomic and Clinical Data • Project GENIE launched with data • Project GENIE aims to promote • Data from Project GENIE may guide from 19,000 patients from 8 institu data sharing to enhance precision identification of drug targets and tions with a variety of tumor types . medicine research . biomarkers in patients with cancer . Genomic profi ling of tumors accessible in the cBioPortal for Cancer Genomics. Despite the has become increasingly com- differences in genomic testing at the contributing centers, the mon across cancer types, but genomic data collected so far are largely concordant across the data are not regularly made the centers, and mutation rates are similar to those reported by available to the entire research The Cancer Genome Atlas. Initial results from Project GENIE community. The American Asso- suggest that more than 30% of tumors harbor potentially ciation for Cancer Research clinically actionable mutations. Advantages of this platform (AACR) launched the Genomics, include integration of clinical data from electronic health Evidence, Neoplasia, Informa- records and increased statistical power to potentially facilitate tion, Exchange (GENIE) project understanding of the clinical relevance of somatic mutations in partnership with eight academic institutions to facilitate and improve patient selection for targeted therapies. The large-scale sharing of genomic and clinical data. The AACR establishment of infrastructure for integrating genomic and Project GENIE Consortium released the fi rst of set of data clinical data has the potential to aid identifi cation of thera- from 19,000 patients in January 2017, and the number of sam- peutic targets and biomarkers of treatment and response in ples included is expected to grow to more than 100,000 within patients with cancer to enhance precision medicine research 5 years as more centers join the Consortium. -

Proquest Dissertations
RICE UNIVERSITY Molecular Basis of Gene Dosage Sensitivity by Jianping Chen A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE Doctor of Philosophy APPROVED, THESIS COMMITTEE: Ariel Fernandez, Chair Karl F. Hasselmann Professor Department of Bioengineering Rice University rn'idtfP h.tQsJUW*- Michael W. Deem, John W. Cox Professor Department of Bioengineering Department of Physics and Astronomy Rice Universit ^engineering at Rice University Jiochemistry at Baylor College of Medicine Axxttig- «J^gb/uv Laura Segaton, T.N. Law Assistant Professor Chemical and Biomolecular Engineering Rice University HOUSTON, TEXAS JANUARY 2009 UMI Number: 3362141 INFORMATION TO USERS The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleed-through, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. UMI® UMI Microform 3362141 Copyright 2009 by ProQuest LLC All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest LLC 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106-1346 ABSTRACT Molecular Basis of Gene Dosage Sensitivity by JianpingChen Deviation of gene expression from normal levels has been associated with diseases. Both under- and overexpression of genes could lead to deleterious biological consequences. Dosage balance has been proposed to be a key issue of determining gene expression pheno- type.