Characterization of the Complete Chloroplast Genome of Platycarya
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

5 Fagaceae Trees
CHAPTER 5 5 Fagaceae Trees Antoine Kremerl, Manuela Casasoli2,Teresa ~arreneche~,Catherine Bod6n2s1, Paul Sisco4,Thomas ~ubisiak~,Marta Scalfi6, Stefano Leonardi6,Erica ~akker~,Joukje ~uiteveld', Jeanne ~omero-Seversong, Kathiravetpillai Arumuganathanlo, Jeremy ~eror~',Caroline scotti-~aintagne", Guy Roussell, Maria Evangelista Bertocchil, Christian kxerl2,Ilga porth13, Fred ~ebard'~,Catherine clark15, John carlson16, Christophe Plomionl, Hans-Peter Koelewijn8, and Fiorella villani17 UMR Biodiversiti Genes & Communautis, INRA, 69 Route d'Arcachon, 33612 Cestas, France, e-mail: [email protected] Dipartimento di Biologia Vegetale, Universita "La Sapienza", Piazza A. Moro 5,00185 Rome, Italy Unite de Recherche sur les Especes Fruitikres et la Vigne, INRA, 71 Avenue Edouard Bourlaux, 33883 Villenave d'Ornon, France The American Chestnut Foundation, One Oak Plaza, Suite 308 Asheville, NC 28801, USA Southern Institute of Forest Genetics, USDA-Forest Service, 23332 Highway 67, Saucier, MS 39574-9344, USA Dipartimento di Scienze Ambientali, Universitk di Parma, Parco Area delle Scienze 1lIA, 43100 Parma, Italy Department of Ecology and Evolution, University of Chicago, 5801 South Ellis Avenue, Chicago, IL 60637, USA Alterra Wageningen UR, Centre for Ecosystem Studies, P.O. Box 47,6700 AA Wageningen, The Netherlands Department of Biological Sciences, University of Notre Dame, Notre Dame, IN 46556, USA lo Flow Cytometry and Imaging Core Laboratory, Benaroya Research Institute at Virginia Mason, 1201 Ninth Avenue, Seattle, WA 98101, -

Synteny Analysis in Rosids with a Walnut Physical Map Reveals Slow Genome Evolution in Long-Lived Woody Perennials Ming-Cheng Luo1, Frank M
Luo et al. BMC Genomics (2015) 16:707 DOI 10.1186/s12864-015-1906-5 RESEARCH ARTICLE Open Access Synteny analysis in Rosids with a walnut physical map reveals slow genome evolution in long-lived woody perennials Ming-Cheng Luo1, Frank M. You2, Pingchuan Li2, Ji-Rui Wang1,4, Tingting Zhu1, Abhaya M. Dandekar1, Charles A. Leslie1, Mallikarjuna Aradhya3, Patrick E. McGuire1 and Jan Dvorak1* Abstract Background: Mutations often accompany DNA replication. Since there may be fewer cell cycles per year in the germlines of long-lived than short-lived angiosperms, the genomes of long-lived angiosperms may be diverging more slowly than those of short-lived angiosperms. Here we test this hypothesis. Results: We first constructed a genetic map for walnut, a woody perennial. All linkage groups were short, and recombination rates were greatly reduced in the centromeric regions. We then used the genetic map to construct a walnut bacterial artificial chromosome (BAC) clone-based physical map, which contained 15,203 exonic BAC-end sequences, and quantified with it synteny between the walnut genome and genomes of three long-lived woody perennials, Vitis vinifera, Populus trichocarpa,andMalus domestica, and three short-lived herbs, Cucumis sativus, Medicago truncatula, and Fragaria vesca. Each measure of synteny we used showed that the genomes of woody perennials were less diverged from the walnut genome than those of herbs. We also estimated the nucleotide substitution rate at silent codon positions in the walnut lineage. It was one-fifth and one-sixth of published nucleotide substitution rates in the Medicago and Arabidopsis lineages, respectively. We uncovered a whole-genome duplication in the walnut lineage, dated it to the neighborhood of the Cretaceous-Tertiary boundary, and allocated the 16 walnut chromosomes into eight homoeologous pairs. -

Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Quercus Bawanglingensis Huang, Li Et Xing, a Vulnerable Oak Tree in China
Article Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Quercus bawanglingensis Huang, Li et Xing, a Vulnerable Oak Tree in China Xue Liu 1 , Er-Mei Chang 1, Jian-Feng Liu 1,* , Yue-Ning Huang 1, Ya Wang 1, Ning Yao 1 and Ze-Ping Jiang 1,2 1 Key Laboratory of Tree Breeding and Cultivation of State Forestry Administration, Research Institute of Forestry, Chinese Academy of Forestry, Beijing 100091, China 2 Research Institute of Forest Ecology, Environment and Protection, Chinese Academy of Forestry, Beijing 100091, China * Correspondence: [email protected] Received: 5 June 2019; Accepted: 12 July 2019; Published: 15 July 2019 Abstract: Quercus bawanglingensis Huang, Li et Xing, an endemic evergreen oak of the genus Quercus (Fagaceae) in China, is currently listed in the Red List of Chinese Plants as a vulnerable (VU) plant. No chloroplast (cp) genome information is currently available for Q. bawanglingensis, which would be essential for the establishment of guidelines for its conservation and breeding. In the present study, the cp genome of Q. bawanglingensis was sequenced and assembled into double-stranded circular DNA with a length of 161,394 bp. Two inverted repeats (IRs) with a total of 51,730 bp were identified, and the rest of the sequence was separated into two single-copy regions, namely, a large single-copy (LSC) region (90,628 bp) and a small single-copy (SSC) region (19,036 bp). The genome of Q. bawanglingensis contains 134 genes (86 protein-coding genes, 40 tRNAs and eight rRNAs). More forward (29) than inverted long repeats (21) are distributed in the cp genome. -

Characterization of the Complete Chloroplast Genome of Platycarya Strobilacea (Juglandaceae)
Conservation Genet Resour (2017) 9:79–81 DOI 10.1007/s12686-016-0624-x TECHNICAL NOTE Characterization of the complete chloroplast genome of Platycarya strobilacea (Juglandaceae) Jing Yan1 · Kai Han1 · Shuyun Zeng1 · Peng Zhao1 · Keith Woeste2 · Jianfang Li1 · Zhan-Lin Liu1 Received: 1 September 2016 / Accepted: 4 October 2016 / Published online: 6 October 2016 © Springer Science+Business Media Dordrecht 2016 Abstract. The whole chloroplast genome (cp genome) The overall AT content of the cp genome is 64 %, and the sequence of Platycarya strobilacea was characterized corresponding values of the LSC, SSC and IR regions are from Illumina pair-end sequencing data. The complete cp 66.4, 70.1 and 57.5 %, respectively. Phylogenetic analysis genome was 160,994 bp in length and contained a large confirmed the placement of P. strobilacea near to Juglans. single copy region (LSC) of 90,225 bp and a small single copy region (SSC) of 18,371 bp, which were separated Keywords Platycarya strobilacea · Hickory · China · by a pair of inverted repeat regions (IRs, 26,199 bp). The Conservation · Chloroplast genome genome contained 130 genes, including 85 protein-coding genes (80 PCG species), 36 tRNA genes (29 tRNA spe- cies) and 8 ribosomal RNA genes (4 rRNA species). Most Platycarya strobilacea, the only species in the monotypic genes occur as a single copy, but 15 genes are duplicated. genus Platycarya (Juglandaceae), mainly grows as scat- tered individuals in evergreen forests of South China (Chen et al. 2012). As one of main components of the local vegeta- tion, this species plays a key role in the forest ecosystems. -

(Castanea) Cultivar 'Yanshanzaofeng'
Advance Journal of Food Science and Technology 5(9): 1192-1197, 2013 ISSN: 2042-4868; e-ISSN: 2042-4876 © Maxwell Scientific Organization, 2013 Submitted: May 20, 2013 Accepted: June 11, 2013 Published: September 05, 2013 A Morphological and Histological Characterization of Male Flower in Chestnut (Castanea) Cultivar ‘Yanshanzaofeng’ Feng Zou, Su-Juan Guo, Huan Xiong, Peng Xie, Wen-Jun Lv and Guang-Hui Li Key Laboratory for Silviculture and Conservation, Ministry of Education, Beijing Forestry University, 100083, Beijing, P.R. China Abstract: Chinese chestnut (Castanea mollissima Blume.) is a widely distributed fruit tree and well known for its ecological and economic value. In order to evaluate obstacles to male reproductive in the C. mollissima, a morphological and histological characterization of male flower of chestnut cultivar ‘Yanshanzaofeng’ were examined by paraffin section technique and scanning electron microscopy. The results showed that male catkins with floral primordia were formed in the buds of one-year olds shoots in later April. Later, a protoderm, ground meristem and a procambium had differentiated in young anthers. Each young anther soon developed to four microsporangia. The anther wall layers developed completely by mid-May and consisted of one-cell-layered epidermis, one-cell-layered endothecium, two or three middle layers and one-cell-layered tapetum. The tapetum was of glandular type. Microspore mother cells underwent meiosis through simultaneous cytokinesis in later May and gave rise to tetrads of microspores, which were tetrahedrally arranged. Mature pollens contained two cells with three germ pores. Anthers were dehiscent and pollen grains shed by early June. Based our results, we did not find the abnormal male flower in the C. -

Wingnut (Juglandaceae)
83 Wingnut (Juglandaceae) as a new generic host for Pityophthorus juglandis (Coleoptera: Curculionidae) and the thousand cankers disease pathogen, Geosmithia morbida (Ascomycota: Hypocreales) Stacy M. Hishinuma, Paul L. Dallara, Mohammad A. Yaghmour, Marcelo M. Zerillo, Corwin M. Parker, Tatiana V. Roubtsova, Tivonne L. Nguyen, Ned A. Tisserat, Richard M. Bostock, Mary L. Flint, Steven J. Seybold1 Abstract—The walnut twig beetle (WTB), Pityophthorus juglandis Blackman (Coleoptera: Curculionidae), vectors a fungus, Geosmithia morbida Kolařík, Freeland, Utley, and Tisserat (Ascomycota: Hypocreales), which colonises and kills the phloem of walnut and butternut trees, Juglans Linnaeus (Juglandaceae). Over the past two decades, this condition, known as thousand cankers disease (TCD), has led to the widespread mortality of Juglans species in the United States of America. Recently the beetle and pathogen were discovered on several Juglans species in northern Italy. Little is known about the extra-generic extent of host acceptability and suitability for the WTB. We report the occurrence of both the WTB and G. morbida in three species of wingnut, Pterocarya fraxinifolia Spach, Pterocarya rhoifolia Siebold and Zuccarini, and Pterocarya stenoptera de Candolle (Juglandaceae) growing in the United States Department of Agriculture-Agricultural Research Service, National Clonal Germplasm Repository collection in northern California (NCGR) and in the Los Angeles County Arboretum and Botanic Garden in southern California, United States of America. In two instances (once in P. stenoptera and once in P. fraxinifolia) teneral (i.e., brood) adult WTB emerged and were collected more than four months after infested branch sections had been collected in the field. Koch’s postulates were satisfied with an isolate of G. -

(Juglandaceae) in the Early Miocene of Panama
CHAPTER 7 FRUITS OF OREOMUNNEA (JUGLANDACEAE) IN THE EARLY MIOCENE OF PANAMA Fabiany Herrera, Steven R. Manchester,* Rebecca Koll, and Carlos Jaramillo ABSTRACT Permineralized fruits of Juglandaceae have been recovered from the Early Miocene Cucaracha Formation exposed in excavations of the Panama Canal. Transverse sections and peels reveal that the nuts are subdivided into eight chambers at the base, and the nutshell and septa are composed mainly of fibers. Together, these features are found only in the Neotropical juglandaceous clade composed of extant Alfaroa Standl. and Oreomunnea Oerst. Based on the small nut size and rela- tively thin wall and septa, these nuts are assigned to Oreomunnea, which is distinguished from extant Alfaroa by its winged fruits adapted for wind dispersal. A. Graham first reported pollen grains of the Alfaroa/Oreomunnea type from the Miocene of Panama. The new fossil fruits of the Cucaracha Formation are the earliest macrofossil record of Oreomunnea, enhancing our under- standing of the phytogeographic history of Juglandaceae in Central America. If this newly de- scribed fossil species, O. grahamii Manch. & Herrera, grew in similar ecological settings to those of its modern counterparts, the fossils may be an indicator of the medium-high paleotopographic relief of Panama during the Early Miocene. Key words: Alfaroa, fossils, fruits, Juglandaceae, Miocene, Oreomunnea. The Juglandaceae have an excellent fossil rec- Neotropical genera Alfaroa Standl. and Oreo- ord in the Northern Hemisphere, well represented munnea Oerst. have mostly eluded detection in by pollen, leaves, wood, catkins, and fruits. This the macrofossil record. record includes most of the extant genera, plus a Ten genera in two subfamilies are recognized few extinct ones in the Paleogene (Manchester, for the Juglandaceae (Manos & Stone, 2001; 1987, 1989; Elliot et al., 2006). -

Inflorescence Dimorphism, Heterodichogamy and Thrips
Annals of Botany 113: 467–476, 2014 doi:10.1093/aob/mct278, available online at www.aob.oxfordjournals.org Inflorescence dimorphism, heterodichogamy and thrips pollination in Platycarya strobilacea (Juglandaceae) Tatsundo Fukuhara* and Shin-ichiro Tokumaru Faculty of Education, Fukuoka University of Education, 1-1 Akama-Bunkyo-machi, Munakata, Fukuoka, Japan * For correspondence. E-mail [email protected] Received: 22 July 2013 Returned for revision: 11 September 2013 Accepted: 14 October 2013 Published electronically: 3 December 2013 † Background and Aims Unlike other taxa in Juglandaceae or in closely related families, which are anemophilous, Platycarya strobilacea has been suggested to be entomophilous. In Juglandaceae, Juglans and Carya show hetero- dichogamy, a reproductive strategy in which two morphs coexist in a population and undergo synchronous reciprocal sex changes. However, there has been no study focusing on heterodichogamy in the other six or seven genera, includ- ing Platycarya. † Methods Inflorescence architecture, sexual expression and pollination biology were examined in a P. strobilacea population in Japan. Flowering phenology was monitored daily for 24 trees in 2008 and 27 in 2009. Flower visitors and inhabitants were recorded or collected from different sexes and stages. † Key results The population of P. strobilacea showed heterodichogamous phenology with protogynous and duodi- chogamous–protandrous morphs. This dimorphism in dichogamy was associated with distinct inflorescence morph- ologies.Thrips pollination was suggested bythe frequent presence of thrips withattached pollen grains,the scarcityof other insect visitors, the synchronicity of thrips number in male spikes with the maturation of female flowers, and morphological characters shared with previously reported thrips-pollinated plants. Male spikes went through two consecutive stages: bright yellow and strong-scented M1 stage, and brownish and little-scented M2 stage. -

Fragrant Annuals Fragrant Annuals
TheThe AmericanAmerican GARDENERGARDENER® TheThe MagazineMagazine ofof thethe AAmericanmerican HorticulturalHorticultural SocietySociety JanuaryJanuary // FebruaryFebruary 20112011 New Plants for 2011 Unusual Trees with Garden Potential The AHS’s River Farm: A Center of Horticulture Fragrant Annuals Legacies assume many forms hether making estate plans, considering W year-end giving, honoring a loved one or planting a tree, the legacies of tomorrow are created today. Please remember the American Horticultural Society when making your estate and charitable giving plans. Together we can leave a legacy of a greener, healthier, more beautiful America. For more information on including the AHS in your estate planning and charitable giving, or to make a gift to honor or remember a loved one, please contact Courtney Capstack at (703) 768-5700 ext. 127. Making America a Nation of Gardeners, a Land of Gardens contents Volume 90, Number 1 . January / February 2011 FEATURES DEPARTMENTS 5 NOTES FROM RIVER FARM 6 MEMBERS’ FORUM 8 NEWS FROM THE AHS 2011 Seed Exchange catalog online for AHS members, new AHS Travel Study Program destinations, AHS forms partnership with Northeast garden symposium, registration open for 10th annual America in Bloom Contest, 2011 EPCOT International Flower & Garden Festival, Colonial Williamsburg Garden Symposium, TGOA-MGCA garden photography competition opens. 40 GARDEN SOLUTIONS Plant expert Scott Aker offers a holistic approach to solving common problems. 42 HOMEGROWN HARVEST page 28 Easy-to-grow parsley. 44 GARDENER’S NOTEBOOK Enlightened ways to NEW PLANTS FOR 2011 BY JANE BERGER 12 control powdery mildew, Edible, compact, upright, and colorful are the themes of this beating bugs with plant year’s new plant introductions. -
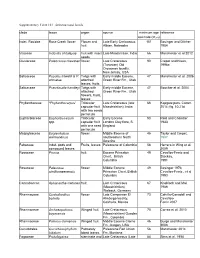
Supplementary Table 10.1. Selected Rosid Fossils Clade Taxon Organ Source Minimum Age Reference Estimate (Mya) Indet
Supplementary Table 10.1. Selected rosid fossils clade taxon organ source minimum age reference estimate (Mya) Indet. Rosidae Rose Creek flower Flower and Late Early Cretaceous 101 Basinger and Dilcher fruit Albian, Nebraska 1984 Vitaceae Indovitis chitaleyae fruit with intact Late Maastrictian, India 66 Manchester et al 2012 seeds Clusiaceae Paleoclusia chevalieri flower Late Cretaceous 90 Crepet and Nixon, (Turonian) Old 1998a Crossman locality, New Jersey, USA Salicaceae Populus tidwellii & P. Twigs with Early middle Eocene, 47 Manchester et al. 2006 wilmatae attached Green River Fm., Utah leaves, fruits Salicaceae Pseudosalix handleyi Twigs with Early middle Eocene, 47 Boucher et al. 2004 attached Green River Fm., Utah flowers, fruits, leaves Phyllanthaceae “Phyllanthocarpus” Trilocular Late Cretaceous (late 66 Kapgate pers. Comm. capsular fruit Maastrichtian), India 2014; fig. 10.21d with two seeds per locule Euphorbiaceae Euphorbiocarpon Trilocular Early Eocene 50 Reid and Chandler spp. capsular fruit London Clay flora, S. 1933 with one seed England per locule Malpighiaceae Eoglandulosa flower Middle Eocene of 45 Taylor and Crepet, warmanensis southeastern North 1987 America Fabaceae Indet. pods and Fruits, leaves Paleocene of Colombia 58 Herrera in Wing et al. compound leaves 2009 Rosaceae Prunus fruit Eocene Princeton 49 Cevallos-Ferriz and Chert, British Stockey, Columbia 1991 Rosaceae Paleorosa flower Middle Eocene 49 Basinger 1976; similkameenensis Princeton Chert, British Cevallos-Ferriz , et al Columbia 1993 Cannabaceae Aphananthe cretacea fruit Late Cretaceous 67 Knobloch and Mai (Maastrichtian) 1986 Walbeck, Germany Rhamnaceae Coahuilanthus flower Late Campanian El 73 Calvillo-Canadell and belinda Almácigo locality, Cevallos- Coahuila, Mexico Ferriz 2007 Rhamnaceae Archaeopaliurus Winged fruit Late Cretaceous 70 Correa et al. -

Multiple Host Shifts Between Distantly Related Plants, Juglandaceae And
Molecular Phylogenetics and Evolution 38 (2006) 231–240 www.elsevier.com/locate/ympev Multiple host shifts between distantly related plants, Juglandaceae and Ericaceae, in the leaf-mining moth Acrocercops leucophaea complex (Lepidoptera: Gracillariidae) Issei Ohshima ¤, Kazunori Yoshizawa Systematic Entomology, Department of Ecology and Systematics, Graduate School of Agriculture, Hokkaido University, Sapporo 060-8589, Japan Received 4 April 2005; revised 8 June 2005 Available online 20 July 2005 Abstract Insect herbivores such as gall formers and leaf miners are often highly specialized and adapted to their respective natal host plants. Due to the specialization and adaptation, it is presumed that host shifts readily occur among closely related plant species. Leaf-mining moths, the Acrocercops leucophaea complex, consist of three species, A. leucophaea, A. deWgurata, and A. transecta. Lar- vae of all the species of the complex feed on Juglandaceae plants, but A. leucophaea and A. transecta are also associated with an Eric- aceae plant, which is quite distantly related to Juglandaceae. Such a host utilization as in this species complex is very rare among phytophagous insects. In the present study, we estimate the history of host shifts by reconstructing the phylogeny of the A. leucop- haea complex using molecular data (partial sequence of mitochondrial COI, 12S rDNA, and ND5). Parsimony and maximum likeli- hood analyses indicated that the common ancestor of the A. leucophaea complex used Juglandaceae only, and that the association with Ericaceae has evolved in A. leucophaea and A. transecta independently. Parametric bootstrap analysis also supported multiple origins of the association with Ericaceae in this complex. These results imply that there are ecological and biochemical factors that promote host shifting between Juglandaceae and Ericaceae despite the two families being not closely related. -

Whole Genome Based Insights Into the Phylogeny and Evolution of the Juglandaceae
Whole Genome based Insights into the Phylogeny and Evolution of the Juglandaceae Huijuan Zhou Northwest A&F University: Northwest Agriculture and Forestry University Yiheng Hu Northwestern University Aziz Ebrahimi Purdue University Peiliang Liu Northwestern University Keith Woeste Purdue University Shuoxin Zhang Northwest A&F University: Northwest Agriculture and Forestry University Peng Zhao ( [email protected] ) Northwest University https://orcid.org/0000-0003-3033-6982 Research article Keywords: Diversication, Divergence time, Genome, Juglandaceae, Phylogenomics, Plastome Posted Date: May 24th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-495294/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/23 Abstract Background: The walnut family (Juglandaceae) contains commercially important woody trees commonly called walnut, wingnut, pecan and hickory. Phylogenetic relationships in the Juglandaceae are problematic, and their historical diversication has not been claried, in part because of low phylogenetic resolution and/or insucient marker variability. Results: We reconstructed the backbone phylogenetic relationships of Juglandaceae using organelle and nuclear genome data from 27 species. The divergence time of Juglandaceae was estimated to be 78.7 Mya. The major lineages diversied in warm and dry habitats during the mid-Paleocene and early Eocene. The plastid, mitochondrial, and nuclear phylogenetic analyses all revealed three subfamilies, i.e., Juglandoideae, Engelhardioideae, Rhoipteleoideae. Five genera of Juglandoideae were strongly supported. Juglandaceae were estimated to have originated during the late Cretaceous, while Juglandoideae were estimated to have originated during the Paleocene, with evidence for rapid diversication events during several glacial and geological periods. The phylogenetic analyses of organelle sequences and nuclear genome yielded highly supported incongruence positions for J.