Seasonal Variability in the Microbial Community and Pathogens in Wastewater Final Effluents
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

Indagine Sulla Presenza Di Helicobacter Pullorum in Allevamenti Avicoli Italiani
Alma Mater Studiorum – Università di Bologna DOTTORATO DI RICERCA IN “EPIDEMIOLOGIA E CONTROLLO DELLE ZOONOSI” Ciclo XX Settore scientifico disciplinare di afferenza: VET-05 TITOLO TESI Indagine sulla presenza di Helicobacter pullorum in allevamenti avicoli italiani Presentata da: Dr. Mirko Rossi Coordinatore Dottorato Relatore Prof.Luigi Morganti Prof. Renato Giulio Zanoni Esame finale anno 2008 1 2 Para a Joana Hà-de haver uma cor por descobrir Um juntar de palava escondido Hà-de haver uma chave para abrir A porta deste muro desmedido (Josè Saramago, Hà-de aver…) 3 4 INDICE PARTE GENERALE 7 Capitolo 1 Genere Helicobacter 9 Capitolo 2 Helicobacter pullorum 21 PARTE SPERIMENTALE 31 Presentazione 33 Capitolo 1 Ottimizzazione dei metodi di isolamento di Helicobacter pullorum da matrice policontaminata 35 Capitolo 2 Isolamento di Helicobacter pullorum da contenuto ciecale di broiler, galline ovaiole, tacchini e struzzo, caratterizzazione fenotipica e tipizzazione genotipica degli isolati 55 Capitolo 3 Indagine sulla presenza di Helicobacter pullorum da feci di pazienti umani affetti da patologia gastroenterica 85 Capitolo 4. Antibiotico resistenza in Helicobacter pullorum 93 Conclusioni 119 RINGRAZIAMENTI 123 BIBLIOGRAFIA 125 5 6 PARTE GENERALE 7 8 CAPITOLO 1 Genere Helicobacter 1.1 Tassonomia del genere Helicobacter Il genere Helicobacter è stato originariamente descritto nel 1989 da Goodwin et al . i quali, sulla base di caratteristiche fenotipiche e genotipiche quali la composizione in acidi grassi della membrana cellulare, la sensibilità ad alcuni antibiotici e la sequenza del rRNA, classificarono in questo nuovo genere le specie [Campylobacter pylori ] e [Campylobacter mustelae ], isolate rispettivamente dallo stomaco di uomo (Marshall et al. , 1985) e di furetto (Fox et al ., 1988). -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

Host-Adaptation in Legionellales Is 2.4 Ga, Coincident with Eukaryogenesis
bioRxiv preprint doi: https://doi.org/10.1101/852004; this version posted February 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Host-adaptation in Legionellales is 2.4 Ga, 2 coincident with eukaryogenesis 3 4 5 Eric Hugoson1,2, Tea Ammunét1 †, and Lionel Guy1* 6 7 1 Department of Medical Biochemistry and Microbiology, Science for Life Laboratories, 8 Uppsala University, Box 582, 75123 Uppsala, Sweden 9 2 Department of Microbial Population Biology, Max Planck Institute for Evolutionary 10 Biology, D-24306 Plön, Germany 11 † current address: Medical Bioinformatics Centre, Turku Bioscience, University of Turku, 12 Tykistökatu 6A, 20520 Turku, Finland 13 * corresponding author 14 1 bioRxiv preprint doi: https://doi.org/10.1101/852004; this version posted February 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 15 Abstract 16 Bacteria adapting to living in a host cell caused the most salient events in the evolution of 17 eukaryotes, namely the seminal fusion with an archaeon 1, and the emergence of both the 18 mitochondrion and the chloroplast 2. A bacterial clade that may hold the key to understanding 19 these events is the deep-branching gammaproteobacterial order Legionellales – containing 20 among others Coxiella and Legionella – of which all known members grow inside eukaryotic 21 cells 3. -

Electronic Laboratory Reporting Use Case January 2011
ILLINOIS HEALTH INFORMATION EXCHANGE Electronic Laboratory Reporting Use Case Electronic Laboratory Reporting and Health Information Exchange Illinois Health Information Exchange Public Health Work Group January 2011 Electronic Laboratory Reporting Use Case January 2011 Table of Contents 1.0 Executive Summary……………………………………………………………………….3 2.0 Introduction…………………………………………………………………...……………..5 3.0 Scope……………………………………..………………………………………………………5 4.0 Use Case Stakeholders…………………………………………………………….….....6 5.0 Issues and Obstacles……………………………………………………………………...8 6.0 Use Case Pre-Conditions .………………….…………………………………………...8 7.0 Use Case Post-Conditions.……………………………………………………………...9 8.0 Detailed Scenarios/Technical Specifications.………………………………10 9.0 Validation and Certification………………………………………………………...12 Appendix ………………………………………………………………………………………….....13 Page 2 Electronic Laboratory Reporting Use Case January 2011 1.0 Executive Summary This Use Case is a product of the Public Health Work Group (PHWG) of the Illinois Health Information Exchange (HIE) Advisory Committee. The Illinois HIE Advisory Committee was constituted as the diverse public healthcare stakeholder body providing input and recommendations on the creation of the Illinois Health Information Exchange Authority (“the Authority”) as the Illinois vehicle for designing and implementing electronic health information exchange in Illinois. The establishment of the Authority marks the formal transition of the work of the HIE Advisory Committee and the Work Groups into alignment with the provisions of Illinois -

University of Nova Gorica Graduate School
UNIVERSITY OF NOVA GORICA GRADUATE SCHOOL THE STUDY OF OPTIMAL TECHNOLOGICAL PROCEDURES OF INTERNAL PLUMBING SYSTEM DISINFECTION FACILITIES IN USE BY THE SENSITIVE HUMAN POPULATIONS MASTER´S THESIS Janez Škarja Mentor: Assis. Prof. Darko Drev Nova Gorica, 2016 UNIVERZA V NOVI GORICI FAKULTETA ZA PODIPLOMSKI ŠTUDIJ RAZISKAVA OPTIMALNIH TEHNOLOŠKIH POSTOPKOV DEZINFEKCIJE INTERNIH VODOVODNIH OMREŽIJ OBJEKTOV, KI JIH UPORABLJA OBČUTLJIVEJŠA POPULACIJA LJUDI MAGISTRSKO DELO Janez Škarja Mentor: doc.dr. Darko Drev Nova Gorica, 2016 SUMMARY In a developed world, water is used in a variety of installations and devices for the improvement of life standard. It is important for these elements to be suitably managed and maintained, otherwise they can present a risk to people's health. Although potable water from a public plumbing system coming via the water supply into an internal plumbing system normally is compliant with the regulations, the quality of water in an internal plumbing system often changes – water gets contaminated. There are several types of microorganisms that can grow in water. While most of them present no threat, there are some that can induce health issues in people. Internal plumbing systems with heated water and the possibility of aerosol releasing present a special concern in this matter. They carry a great potential for the growth and proliferation of bacteria from the genus Legionella that cause Legionnaires' disease and Pontiac Fever. Even the fact that Legionella infection has for several years been mentioned in the accident insurance conditions of insurance companies in relation to receiving the insurance fee as compensation for bed day, shows the extent and foremost the seriousness of the disease (5–15% mortality rate). -

Helicobacter Pylori: Comparative Genomics and Structure-Function Analysis of the Flagellum Biogenesis Protein HP0958
UCC Library and UCC researchers have made this item openly available. Please let us know how this has helped you. Thanks! Title Helicobacter pylori: comparative genomics and structure-function analysis of the flagellum biogenesis protein HP0958 Author(s) de Lacy Clancy, Ceara A. Publication date 2014 Original citation de Lacy Clancy, C. A. 2014. Helicobacter pylori: comparative genomics and structure-function analysis of the flagellum biogenesis protein HP0958. PhD Thesis, University College Cork. Type of publication Doctoral thesis Rights © 2014, Ceara A. De Lacy Clancy. http://creativecommons.org/licenses/by-nc-nd/3.0/ Item downloaded http://hdl.handle.net/10468/1684 from Downloaded on 2021-10-10T14:33:51Z Helicobacter pylori: Comparative genomics and structure-function analysis of the flagellum biogenesis protein HP0958 A Thesis Presented in Partial Fulfilment of the Requirements for the Degree of Doctor of Philosophy by Ceara de Lacy Clancy, B.Sc. School of Microbiology National University of Ireland, Cork Supervisor: Prof. Paul W. O’Toole Head of School of Microbiology: Prof. Gerald Fitzgerald February 2014 Table of Contents Table of Contents Abstract ........................................................................................................................ i Chapter 1 Literature Review....................................................................................................... 1 1 Helicobacter pylori .................................................................................................. 2 1.1 Discovery -
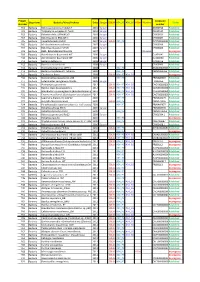
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

EVALUATION of the P45 MOBILE INTEGRATIVE ELEMENT and ITS ROLE IN
EVALUATION OF THE p45 MOBILE INTEGRATIVE ELEMENT AND ITS ROLE IN Legionella pneumophila VIRULENCE A Dissertation by LANETTE M. CHRISTENSEN Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Jeffrey D. Cirillo Committee Members, James Samuel Jon Skare Farida Sohrabji Head of Program, Warren Zimmer May 2018 Major Subject: Medical Sciences Copyright 2018 Lanette Christensen ABSTRACT Legionella pneumophila are aqueous environmental bacilli that live within protozoal species and cause a potentially fatal form of pneumonia called Legionnaires’ disease. Not all L. pneumophila strains have the same capacity to cause disease in humans. The majority of strains that cause clinically relevant Legionnaires’ disease harbor the p45 mobile integrative genomic element. Contribution of the p45 element to L. pneumophila virulence and ability to withstand environmental stress were addressed in this study. The L. pneumophila Philadelphia-1 (Phil-1) mobile integrative element, p45, was transferred into the attenuated strain Lp01 via conjugation, designating p45 an integrative conjugative element (ICE). The resulting trans-conjugate, Lp01+p45, was compared with strains Phil-1 and Lp01 to assess p45 in virulence using a guinea pig model infected via aerosol. The p45 element partially recovered the loss of virulence in Lp01 compared to that of Phil-1 evident in morbidity, mortality, and bacterial burden in the lungs at the time of death. This phenotype was accompanied by enhanced expression of type II interferon in the lungs and spleens 48 hours after infection, independent of bacterial burden. -

Bacterial Species Exclusively Identified in Male Subjects
Supplementary Table 1: Bacterial species exclusively identified in male subjects Acholeplasma ales Clostridium magnum Lactobacillus ruminis Pseudomonas collierea Acholeplasma cavigenitalium Clostridium malenominatum Lactobacillus salivarius Pseudomonas jinjuensis Acholeplasma equifetale Clostridium nitrophenolicum Lactobacillus versmoldensis Pseudomonas luteola Acholeplasma granularum Clostridium paraputrificum Lactococcus lactis Pseudomonas panipatensis Acholeplasma hippikon Clostridium perfringens Legionella fallonii Pseudomonas poae Acidaminobacter hydrogenoformans Clostridium proteolyticum Legionella taurinensis Pseudomonas tropicalis Acidaminococcus intestini Clostridium proteolyticus Legionella worsleiensis Pseudonocardia acaciae Clostridium Acidiphilium acidophilum saccharoperbutylacetonicum Lentzea californiensis Pseudonocardia asaccharolytica Pseudonocardia Acidiphilium organovorum Clostridium sulfidigenes Leptotrichia buccalis hydrocarbonoxydans Acidiphilium symbioticum Clostridium termitidis Leptotrichia shahii Pseudoxanthomonas mexicana Acidisoma tundrae Clostridium tetani Leptotrichia wadei Pullulanibacillus naganoensis Acidovorax facilis Clostridium thermoalcaliphilum Leucobacter albus Rhizobium mesoamericanum Acinetobacter indicus Clostridium tunisiense Lewinella marina Rhodanobacter thiooxydans Acinetobacter venetianus Cohnella fontinalis Luteimonas aquatica Rhodobacter blasticus Actinoallomurus luridus Cohnella laeviribosi Luteimonas mephitis Rhodobacter gluconicum Actinobacillus pleuropneumoniae Collinsella stercoris -

Diversity of Mat-Forming Sulfide-Oxidizing Bacteria at Continental Margins
Diversity of Mat-forming Sulfide-oxidizing Bacteria at Continental Margins Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften - Dr. rer. nat. - dem Fachbereich Biologie/Chemie der Universität Bremen vorgelegt von Stefanie Grünke Bremen, April 2010 Die vorliegende Doktorarbeit wurde in der Zeit von Juni 2006 bis April 2010 am Max- Planck-Institut für Marine Mikrobiologie und am Alfred-Wegener-Institut für Polar- und Meeresforschung angefertigt. 1. Gutachterin: Prof. Dr. Antje Boetius 2. Gutachter: Prof. Dr. Rudolf Amann Tag des Promotionskolloquiums: 4. Juni 2010 Diese Arbeit ist all denjenigen gewidmet, die ihre Segel setzen, um neue Welten gu erkunden. Seien sie sich gewiss, dass auf Sturm immer ruhiges Wasserfolgt. Wertrauen sie auf ihr größtes Gut — ihre Freunde und Familie. Nutgen sie ihre Schwächen, um neue Stärken gu finden. Soll Zuversicht ihr Kompass sein! Summary In the oceans, microbial mats formed by chemosynthetic sulfide-oxidizing bacteria are mostly found in so-called ‘reduced habitats’ that are characterized by chemoclines where energy-rich, reduced substances, like hydrogen sulfide, are transported into oxic or suboxic zones. There, these organisms often thrive in narrow zones or gradients of their electron donor (sulfide) and their electron acceptor (mostly oxygen or nitrate). Through the build up of large biomasses, mat-forming sulfide oxidizers may significantly contribute to primary production in their habitats and dense mats represent efficient benthic filters against the toxic gas hydrogen sulfide. As gradient organisms, these mat-forming sulfide oxidizers seem to be adapted to very defined ecological niches with respect to oxygen (or nitrate) and sulfide gradients. However, many aspects regarding their diversity as well as their geological drivers in marine sulfidic habitats required further investigation. -

Three Unusual New Legionella Species
International Journal of Systematic and Evolutionary Microbiology (2001), 51, 1151–1160 Printed in Great Britain Legionella drozanskii sp. nov., Legionella rowbothamii sp. nov. and Legionella fallonii sp. nov.: three unusual new Legionella species 1 Division of Life Sciences, Adenike A. Adeleke,1 Barry S. Fields,2 Robert F. Benson,2 King’s College London, 2 2 3 Franklin-Wilkins Building, Maryam I. Daneshvar, Janet M. Pruckler, Rodney M. Ratcliff, 150 Stamford Street, Timothy G. Harrison,4 Robbin S. Weyant,2 Richard J. Birtles,5 London SE1 8WA, UK Didier Raoult6 and Mahmoud A. Halablab1 2 Division of Bacterial and Mycotic Diseases, Centers for Disease Control and Author for correspondence: Mahmoud A. Halablab. Tel: j44 207 848 4281. Fax: j44 207 848 4500. Prevention (CDC), Atlanta, e-mail: mahmoud.halablab!kcl.ac.uk GA 30333, USA 3 Infectious Diseases Laboratories, Institute of Seven strains of Legionella-like amoebal pathogens (LLAPs) were characterized Medical and Veterinary on the basis of their cultural and staining characteristics, biochemical Science, Frome Road, reactions, serology, cellular fatty acids (CFAs), isoprenoid quinone Adelaide, SA, 5000, Australia composition, total DNA relatedness, analysis of 16S rRNA and macrophage infectivity potentiator (mip) gene sequence analyses. All seven strains 4 PHLS Central Public Health Laboratory, Colindale, exhibited limited growth on buffered charcoal yeast extract α (BCYE) agar, London NW9 5HT, UK required cysteine for growth and contained branched-chain CFAs and quinones 5 Department of typical of Legionella species. The bacilli were Gram-negative and catalase- Microbiology, University of positive. There were varying degrees of serological cross-reactions between Bristol, Bristol BS8 1TD, UK these LLAP strains and other previously described Legionella species.