Comparative Analyses of Legionella Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NCTC) Bacterial Strain Equivalents to American Type Culture Collection (ATCC) Bacterial Strains
This list shows National Collection of Type Cultures (NCTC) bacterial strain equivalents to American Type Culture Collection (ATCC) bacterial strains. NCTC Number CurrentName ATCC Number NCTC 7212 Acetobacter pasteurianus ATCC 23761 NCTC 10138 Acholeplasma axanthum ATCC 25176 NCTC 10171 Acholeplasma equifetale ATCC 29724 NCTC 10128 Acholeplasma granularum ATCC 19168 NCTC 10172 Acholeplasma hippikon ATCC 29725 NCTC 10116 Acholeplasma laidlawii ATCC 23206 NCTC 10134 Acholeplasma modicum ATCC 29102 NCTC 10188 Acholeplasma morum ATCC 33211 NCTC 10150 Acholeplasma oculi ATCC 27350 NCTC 10198 Acholeplasma parvum ATCC 29892 NCTC 8582 Achromobacter denitrificans ATCC 15173 NCTC 10309 Achromobacter metalcaligenes ATCC 17910 NCTC 10807 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 27061 NCTC 10808 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 17062 NCTC 10809 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 27063 NCTC 12156 Acinetobacter baumannii ATCC 19606 NCTC 10303 Acinetobacter baumannii ATCC 17904 NCTC 7844 Acinetobacter calcoaceticus ATCC 15308 NCTC 12983 Acinetobacter calcoaceticus ATCC 23055 NCTC 8102 acinetobacter dna group 13 ATCC 17903 NCTC 10304 Acinetobacter genospecies 13 ATCC 17905 NCTC 10306 Acinetobacter haemolyticus ATCC 17907 NCTC 10305 Acinetobacter haemolyticus subsp haemolyticus ATCC 17906 NCTC 10308 Acinetobacter johnsonii ATCC 17909 NCTC 10307 Acinetobacter junii ATCC 17908 NCTC 5866 Acinetobacter lwoffii ATCC 15309 NCTC 12870 Actinobacillus delphinicola ATCC 700179 NCTC 8529 Actinobacillus equuli ATCC 19392 -

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

Host-Adaptation in Legionellales Is 2.4 Ga, Coincident with Eukaryogenesis
bioRxiv preprint doi: https://doi.org/10.1101/852004; this version posted February 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Host-adaptation in Legionellales is 2.4 Ga, 2 coincident with eukaryogenesis 3 4 5 Eric Hugoson1,2, Tea Ammunét1 †, and Lionel Guy1* 6 7 1 Department of Medical Biochemistry and Microbiology, Science for Life Laboratories, 8 Uppsala University, Box 582, 75123 Uppsala, Sweden 9 2 Department of Microbial Population Biology, Max Planck Institute for Evolutionary 10 Biology, D-24306 Plön, Germany 11 † current address: Medical Bioinformatics Centre, Turku Bioscience, University of Turku, 12 Tykistökatu 6A, 20520 Turku, Finland 13 * corresponding author 14 1 bioRxiv preprint doi: https://doi.org/10.1101/852004; this version posted February 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 15 Abstract 16 Bacteria adapting to living in a host cell caused the most salient events in the evolution of 17 eukaryotes, namely the seminal fusion with an archaeon 1, and the emergence of both the 18 mitochondrion and the chloroplast 2. A bacterial clade that may hold the key to understanding 19 these events is the deep-branching gammaproteobacterial order Legionellales – containing 20 among others Coxiella and Legionella – of which all known members grow inside eukaryotic 21 cells 3. -

Genetic and Functional Studies of the Mip Protein of Legionella
t1.ì. Genetic and Functional Studies of the Mip Protein of Legionella Rodney Mark Ratcliff, BSc (Hons)' MASM Infectious Diseases Laboratories Institute of Medical and Veterinary Science and Department of Microbiology and Immunology UniversitY of Adelaide. Adelaide, South Australia A thesis submitted to the University of Adelaide for the degree of I)octor of Philosophy 15'h March 2000 amended 14th June 2000 Colonies of several Legionella strains on charcoal yeast extract agar (CYE) after 4 days incubation at 37"C in air. Various magnifications show typical ground-glass opalescent appearance. Some pure strains exhibit pleomorphic growth or colour. The top two photographs demonstrate typical red (LH) and blue-white (RH) fluorescence exhibited by some species when illuminated by a Woods (IJV) Lamp. * t Table of Contents .1 Chapter One: Introduction .1 Background .............'. .2 Morphology and TaxonomY J Legionellosis ............. 5 Mode of transmission "..'....'. 7 Environmental habitat 8 Interactions between Legionella and phagocytic hosts 9 Attachment 11 Engulfment and internalisation.'.. 13 Intracellular processing 13 Intracellular replication of legionellae .. " "' " "' 15 Host cell death and bacterial release 18 Virulence (the Genetic factors involved with intracellular multiplication and host cell killing .20 icm/dot system) Legiolysin .25 Msp (Znn* metaloprotease) ...'..... .25 .28 Lipopolysaccharide .29 The association of flagella with disease.. .30 Type IV fimbriae.... .31 Major outer membrane proteins....'.......'. JJ Heat shock proteins'.'. .34 Macrophage infectivity potentiator (Mip) protein Virulenceiraits of Legionella species other than L. pneumophila..........' .39 phylogeny .41 Chapter One (continued): Introduction to bacterial classification and .41 Identificati on of Legionella...'.,..'.. .46 Phylogeny .52 Methods of phylogenetic analysis' .53 Parsimony methods.'.. .55 Distance methods UPGMA cluster analYsis.'.'... -
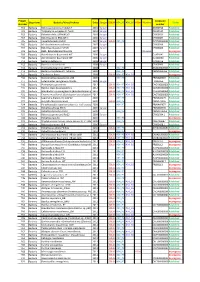
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

EVALUATION of the P45 MOBILE INTEGRATIVE ELEMENT and ITS ROLE IN
EVALUATION OF THE p45 MOBILE INTEGRATIVE ELEMENT AND ITS ROLE IN Legionella pneumophila VIRULENCE A Dissertation by LANETTE M. CHRISTENSEN Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Jeffrey D. Cirillo Committee Members, James Samuel Jon Skare Farida Sohrabji Head of Program, Warren Zimmer May 2018 Major Subject: Medical Sciences Copyright 2018 Lanette Christensen ABSTRACT Legionella pneumophila are aqueous environmental bacilli that live within protozoal species and cause a potentially fatal form of pneumonia called Legionnaires’ disease. Not all L. pneumophila strains have the same capacity to cause disease in humans. The majority of strains that cause clinically relevant Legionnaires’ disease harbor the p45 mobile integrative genomic element. Contribution of the p45 element to L. pneumophila virulence and ability to withstand environmental stress were addressed in this study. The L. pneumophila Philadelphia-1 (Phil-1) mobile integrative element, p45, was transferred into the attenuated strain Lp01 via conjugation, designating p45 an integrative conjugative element (ICE). The resulting trans-conjugate, Lp01+p45, was compared with strains Phil-1 and Lp01 to assess p45 in virulence using a guinea pig model infected via aerosol. The p45 element partially recovered the loss of virulence in Lp01 compared to that of Phil-1 evident in morbidity, mortality, and bacterial burden in the lungs at the time of death. This phenotype was accompanied by enhanced expression of type II interferon in the lungs and spleens 48 hours after infection, independent of bacterial burden. -

Immunoproteomic Identification of Biomarkers for Diagnosis of Legionellosis
Immunoproteomic identification of biomarkers for diagnosis of legionellosis Submitted in total fulfilment of the requirements for the degree of Doctor of philosophy by Kaylass Poorun Department of Chemistry and Biotechnology Faculty of Science, Engineering and Technology Swinburne University of Technology Australia 2014 Abstract Abstract Legionellosis, a disease with significant mortality and morbidity rates, is considered to be the second most frequent cause of severe community-acquired pneumonia. It is difficult to distinguish from other types of pneumonia due to similar clinical manifestations. Several studies have demonstrated the inadequacies of current diagnostic tests for confirming Legionella infections. This study was aimed at identifying biomarkers that can be used in an improved test. A comparative proteomic analysis, using DIGE, was carried out between L. pneumophila ATCC33152 and L. longbeachae NSW150 and D4968 isolates. While many homologous proteins were found to be commonly expressed, numerous others were identified to be differentially expressed under similar in vitro conditions suggesting that the two species have different lifestyles and infection strategies. The bacterial immunoglobulin domain containing protein, found to share sequence homology to Type V secretion proteins intimin and invasin, is not known to be present in Legionella. Human sera containing antibodies against Legionella from a set of blind samples were identified by ELISA. Downstream analyses revealed that diverse immunogens may be responsible for eliciting immune response in different Legionella species which in turn show little to no congeneric cross-reactivity. To the best of our knowledge, this is a unique finding not previously reported. Several serological diagnostic tests currently in use do not include many Legionella species in their testing panel, which may be a reason for many Legionella species being under-reported. -

Bacterial Species Exclusively Identified in Male Subjects
Supplementary Table 1: Bacterial species exclusively identified in male subjects Acholeplasma ales Clostridium magnum Lactobacillus ruminis Pseudomonas collierea Acholeplasma cavigenitalium Clostridium malenominatum Lactobacillus salivarius Pseudomonas jinjuensis Acholeplasma equifetale Clostridium nitrophenolicum Lactobacillus versmoldensis Pseudomonas luteola Acholeplasma granularum Clostridium paraputrificum Lactococcus lactis Pseudomonas panipatensis Acholeplasma hippikon Clostridium perfringens Legionella fallonii Pseudomonas poae Acidaminobacter hydrogenoformans Clostridium proteolyticum Legionella taurinensis Pseudomonas tropicalis Acidaminococcus intestini Clostridium proteolyticus Legionella worsleiensis Pseudonocardia acaciae Clostridium Acidiphilium acidophilum saccharoperbutylacetonicum Lentzea californiensis Pseudonocardia asaccharolytica Pseudonocardia Acidiphilium organovorum Clostridium sulfidigenes Leptotrichia buccalis hydrocarbonoxydans Acidiphilium symbioticum Clostridium termitidis Leptotrichia shahii Pseudoxanthomonas mexicana Acidisoma tundrae Clostridium tetani Leptotrichia wadei Pullulanibacillus naganoensis Acidovorax facilis Clostridium thermoalcaliphilum Leucobacter albus Rhizobium mesoamericanum Acinetobacter indicus Clostridium tunisiense Lewinella marina Rhodanobacter thiooxydans Acinetobacter venetianus Cohnella fontinalis Luteimonas aquatica Rhodobacter blasticus Actinoallomurus luridus Cohnella laeviribosi Luteimonas mephitis Rhodobacter gluconicum Actinobacillus pleuropneumoniae Collinsella stercoris -

Three Unusual New Legionella Species
International Journal of Systematic and Evolutionary Microbiology (2001), 51, 1151–1160 Printed in Great Britain Legionella drozanskii sp. nov., Legionella rowbothamii sp. nov. and Legionella fallonii sp. nov.: three unusual new Legionella species 1 Division of Life Sciences, Adenike A. Adeleke,1 Barry S. Fields,2 Robert F. Benson,2 King’s College London, 2 2 3 Franklin-Wilkins Building, Maryam I. Daneshvar, Janet M. Pruckler, Rodney M. Ratcliff, 150 Stamford Street, Timothy G. Harrison,4 Robbin S. Weyant,2 Richard J. Birtles,5 London SE1 8WA, UK Didier Raoult6 and Mahmoud A. Halablab1 2 Division of Bacterial and Mycotic Diseases, Centers for Disease Control and Author for correspondence: Mahmoud A. Halablab. Tel: j44 207 848 4281. Fax: j44 207 848 4500. Prevention (CDC), Atlanta, e-mail: mahmoud.halablab!kcl.ac.uk GA 30333, USA 3 Infectious Diseases Laboratories, Institute of Seven strains of Legionella-like amoebal pathogens (LLAPs) were characterized Medical and Veterinary on the basis of their cultural and staining characteristics, biochemical Science, Frome Road, reactions, serology, cellular fatty acids (CFAs), isoprenoid quinone Adelaide, SA, 5000, Australia composition, total DNA relatedness, analysis of 16S rRNA and macrophage infectivity potentiator (mip) gene sequence analyses. All seven strains 4 PHLS Central Public Health Laboratory, Colindale, exhibited limited growth on buffered charcoal yeast extract α (BCYE) agar, London NW9 5HT, UK required cysteine for growth and contained branched-chain CFAs and quinones 5 Department of typical of Legionella species. The bacilli were Gram-negative and catalase- Microbiology, University of positive. There were varying degrees of serological cross-reactions between Bristol, Bristol BS8 1TD, UK these LLAP strains and other previously described Legionella species. -

Comparative Analyses of Legionella Species Identifies Genetic Features
Gomez-Valero et al. Genome Biology 2014, 15:505 http://genomebiology.com/2014/15/11/505 RESEARCH Open Access Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires’ disease Laura Gomez-Valero1,2*, Christophe Rusniok1,2, Monica Rolando1,2, Mario Neou1,2, Delphine Dervins-Ravault1,2, Jasmin Demirtas1,2, Zoe Rouy9, Robert J Moore10, Honglei Chen10, Nicola K Petty3, Sophie Jarraud4,5, Jerome Etienne4,5, Michael Steinert6, Klaus Heuner7, Simonetta Gribaldo8, Claudine Médigue9, Gernot Glöckner11, Elizabeth L Hartland12 and Carmen Buchrieser1* Abstract Background: The genus Legionella comprises over 60 species. However, L. pneumophila and L. longbeachae alone cause over 95% of Legionnaires’ disease. To identify the genetic bases underlying the different capacities to cause disease we sequenced and compared the genomes of L. micdadei, L. hackeliae and L. fallonii (LLAP10), which are all rarely isolated from humans. Results: We show that these Legionella species possess different virulence capacities in amoeba and macrophages, correlating with their occurrence in humans. Our comparative analysis of 11 Legionella genomes belonging to five species reveals highly heterogeneous genome content with over 60% representing species-specific genes; these comprise a complete prophage in L. micdadei, the first ever identified in a Legionella genome. Mobile elements are abundant in Legionella genomes; many encode type IV secretion systems for conjugative transfer, pointing to their importance for adaptation of the genus. The Dot/Icm secretion system is conserved, although the core set of substrates is small, as only 24 out of over 300 described Dot/Icm effector genes are present in all Legionella species. We also identified new eukaryotic motifs including thaumatin, synaptobrevin or clathrin/coatomer adaptine like domains. -

HL7 Implementation Guide for CDA Release 2: NHSN Healthcare Associated Infection (HAI) Reports, Release 2 (U.S
CDAR2L3_IG_HAIRPT_R2_D2_2009FEB HL7 Implementation Guide for CDA Release 2: NHSN Healthcare Associated Infection (HAI) Reports, Release 2 (U.S. Realm) Draft Standard for Trial Use February 2009 © 2009 Health Level Seven, Inc. Ann Arbor, MI All rights reserved HL7 Implementation Guide for CDA R2 NHSN HAI Reports Page 1 Draft Standard for Trial Use Release 2 February 2009 © 2009 Health Level Seven, Inc. All rights reserved. Co-Editor: Marla Albitz Contractor to CDC, Lockheed Martin [email protected] Co-Editor/Co-Chair: Liora Alschuler Alschuler Associates, LLC [email protected] Co-Chair: Calvin Beebe Mayo Clinic [email protected] Co-Chair: Keith W. Boone GE Healthcare [email protected] Co-Editor/Co-Chair: Robert H. Dolin, M.D. Semantically Yours, LLC [email protected] Co-Editor Jonathan Edwards CDC [email protected] Co-Editor Pavla Frazier RN, MSN, MBA Contractor to CDC, Frazier Consulting [email protected] Co-Editor: Sundak Ganesan, M.D. SAIC Consultant to CDC/NCPHI [email protected] Co-Editor: Rick Geimer Alschuler Associates, LLC [email protected] Co-Editor: Gay Giannone M.S.N., R.N. Alschuler Associates LLC [email protected] Co-Editor Austin Kreisler Consultant to CDC/NCPHI [email protected] Primary Editor: Kate Hamilton Alschuler Associates, LLC [email protected] Primary Editor: Brett Marquard Alschuler Associates, LLC [email protected] Co-Editor: Daniel Pollock, M.D. CDC [email protected] HL7 Implementation Guide for CDA R2 NHSN HAI Reports Page 2 Draft Standard for Trial Use Release 2 February 2009 © 2009 Health Level Seven, Inc. All rights reserved. Acknowledgments This DSTU was produced and developed in conjunction with the Division of Healthcare Quality Promotion, National Center for Preparedness, Detection, and Control of Infectious Diseases (NCPDCID), and Centers for Disease Control and Prevention (CDC).