LINC00116 Codes for a Mitochondrial Peptide Linking Respiration and Lipid Metabolism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diapositiva 1
ns 10 A Unmethylated C Methylated ) 8 Normal Normal Tissue Cervix Esophagus 4 cervical esophageal a.u. * *** * *** ( 6 TargetID MAPINFO Position Ca-Ski HeLa SW756 tissue COLO-680N KYSE-180 OACM 5.1C tissue cg04255070 22851591 TSS1500 0.896 0.070 0.082 0.060 0.613 0.622 0.075 0.020 2 (log2) 4 cg01593340 22851587 TSS1500 0.826 0.055 0.051 0.110 0.518 0.518 0.034 0.040 cg16113692 22851502 TSS200 0.964 0.073 0.072 0.030 0.625 0.785 0.018 0.010 2 0 cg26918510 22851422 TSS200 0.985 0.019 0.000 0.000 0.711 0.961 0.010 0.060 expression cg26821579 22851416 TSS200 0.988 0.083 0.058 0.000 0.387 0.955 0.021 0.050 TRMT12 mRNA Expression mRNA TRMT12 0 cg01129966 22851401 TSS200 0.933 0.039 0.077 0.020 0.815 0.963 0.078 0.010 -2 cg25421615 22851383 TSS200 0.958 0.033 0.006 0.000 0.447 0.971 0.037 0.020 mRNA cg17953636 22851321 1stExon 0.816 0.092 0.099 0.080 0.889 0.750 0.136 0.030 -4 Methylated SVIP Unmethylated SVIP relative expression (a.u.) expression relative SVIP D Cervix Esophagus HNC Meth ) HNC Unmeth Cervix Meth 3.0 *** Cervix Unmeth *** Esophagus Meth a.u. ( 2.5 CaCaEsophagus-Ski-Ski Unmeth COLO-680N Haematological Meth TSS B TSS Haematological Unmeth 2.0 -244 bp +104 bp 244 bp +104 bp 1.5 ** * expression -SVIP 1.0 HeLa KYSE-180 HeLa TSS TSS 0.5 -ACTB 244 bp +104 bp - 244 bp +104 bp mRNA 0.0 HeLa SW756 Ca-Ski SW756 SW756 OACM 5.1C TSS KYSE-180 TSS COLO-680NOACM 5.1 C - 244 bp +104 bp 244 bp +104 bp E ) 3.0 * Ca-Ski COLO-680N WT TSS TSS 2.0 a.u. -

Altered Expression and Function of Mitochondrial Я-Oxidation Enzymes
0031-3998/01/5001-0083 PEDIATRIC RESEARCH Vol. 50, No. 1, 2001 Copyright © 2001 International Pediatric Research Foundation, Inc. Printed in U.S.A. Altered Expression and Function of Mitochondrial -Oxidation Enzymes in Juvenile Intrauterine-Growth-Retarded Rat Skeletal Muscle ROBERT H. LANE, DAVID E. KELLEY, VLADIMIR H. RITOV, ANNA E. TSIRKA, AND ELISA M. GRUETZMACHER Department of Pediatrics, UCLA School of Medicine, Mattel Children’s Hospital at UCLA, Los Angeles, California 90095, U.S.A. [R.H.L.]; and Departments of Internal Medicine [D.E.K., V.H.R.] and Pediatrics [R.H.L., A.E.T., E.M.G.], University of Pittsburgh School of Medicine, Magee-Womens Research Institute, Pittsburgh, Pennsylvania 15213, U.S.A. ABSTRACT Uteroplacental insufficiency and subsequent intrauterine creased in IUGR skeletal muscle mitochondria, and isocitrate growth retardation (IUGR) affects postnatal metabolism. In ju- dehydrogenase activity was unchanged. Interestingly, skeletal venile rats, IUGR alters skeletal muscle mitochondrial gene muscle triglycerides were significantly increased in IUGR skel- expression and reduces mitochondrial NADϩ/NADH ratios, both etal muscle. We conclude that uteroplacental insufficiency alters of which affect -oxidation flux. We therefore hypothesized that IUGR skeletal muscle mitochondrial lipid metabolism, and we gene expression and function of mitochondrial -oxidation en- speculate that the changes observed in this study play a role in zymes would be altered in juvenile IUGR skeletal muscle. To test the long-term morbidity associated with IUGR. (Pediatr Res 50: this hypothesis, mRNA levels of five key mitochondrial enzymes 83–90, 2001) (carnitine palmitoyltransferase I, trifunctional protein of -oxi- dation, uncoupling protein-3, isocitrate dehydrogenase, and mi- Abbreviations tochondrial malate dehydrogenase) and intramuscular triglycer- CPTI, carnitine palmitoyltransferase I ides were quantified in 21-d-old (preweaning) IUGR and control IUGR, intrauterine growth retardation rat skeletal muscle. -

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

Role of De Novo Cholesterol Synthesis Enzymes in Cancer Jie Yang1,2, Lihua Wang1,2, Renbing Jia1,2
Journal of Cancer 2020, Vol. 11 1761 Ivyspring International Publisher Journal of Cancer 2020; 11(7): 1761-1767. doi: 10.7150/jca.38598 Review Role of de novo cholesterol synthesis enzymes in cancer Jie Yang1,2, Lihua Wang1,2, Renbing Jia1,2 1. Department of Ophthalmology, Ninth People’s Hospital of Shanghai, Shanghai Jiao Tong University School of Medicine, Shanghai, China. 2. Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai, China. Corresponding authors: Renbing Jia, [email protected] and Lihua Wang, [email protected]. Department of Ophthalmology, Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, No. 639 Zhi Zao Ju Road, Shanghai 200011, China © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See http://ivyspring.com/terms for full terms and conditions. Received: 2019.07.21; Accepted: 2019.11.30; Published: 2020.01.17 Abstract Despite extensive research in the cancer field, cancer remains one of the most prevalent diseases. There is an urgent need to identify specific targets that are safe and effective for the treatment of cancer. In recent years, cancer metabolism has come into the spotlight in cancer research. Lipid metabolism, especially cholesterol metabolism, plays a critical role in membrane synthesis as well as lipid signaling in cancer. This review focuses on the contribution of the de novo cholesterol synthesis pathway to tumorigenesis, cancer progression and metastasis. In conclusion, cholesterol metabolism could be an effective target for novel anticancer treatment. Key words: metabolic reprogramming, de novo cholesterol synthesis, cancer progress Introduction Over the past few decades, numerous published that cholesterol plays a critical role in cancer studies have focused on cancer cell metabolism and progression15-19. -

RESEARCH COMMUNICATION HADHA Is a Potential Predictor Of
HADHA is a Potential Predictor of the Response to Platinum-based Chemotherapy RESEARCH COMMUNICATION HADHA is a Potential Predictor of Response to Platinum-based Chemotherapy for Lung Cancer Taihei Kageyama1, Ryo Nagashio1, 2, Shinichiro Ryuge 3, Toshihide Matsumoto1,5, Akira Iyoda4, Yukitoshi Satoh4, Noriyuki Masuda3, Shi-Xu Jiang5, Makoto Saegusa5, Yuichi Sato1, 2* Abstract To identify a cisplatin resistance predictor to reduce or prevent unnecessary side effects, we firstly established four cisplatin-resistant sub-lines and compared their protein profiles with cisplatin-sensitive parent lung cancer cell lines using two-dimensional gel electrophoresis. Between the cisplatin-resistant and -sensitive cells, a total of 359 protein spots were differently expressed (>1.5 fold), and 217 proteins (83.0%) were identified. We focused on a mitochondrial protein, hydroxyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase alpha subunit (HADHA), which was increased in all cisplatin-resistant cells. Furthermore, pre- treated biopsy specimens taken from patients who showed resistance to platinum-based treatment showed a significantly higher positive rate for HADHA in all cases (p=0.00367), including non-small cell lung carcinomas (p=0.002), small-cell lung carcinomas (p=0.038), and adenocarcinomas (p=0.008). These results suggest that the expression of HADHA may be a useful marker to predict resistance to platinum-based chemotherapy in patients with lung cancer. Keywords: Cisplatin - HADHA - lung cancer - two-dimensional gel electrophoresis Asian Pacific J Cancer Prev, 12, 3457-3463 Introduction cisplatin resistance rose due to a decrease of blood flow in the tumor and increased DNA repair (Stewart, 2007), Lung cancer is the leading cause of cancer-related the mechanisms underlying cisplatin resistance have not death in the world, and the five-year overall survival rate yet been clarified, and an effective cisplatin resistance is still below 16% (Jemal et al., 2009). -

(LCHAD) Deficiency / Mitochondrial Trifunctional Protein (MTF) Deficiency
Long chain acyl-CoA dehydrogenase (LCHAD) deficiency / Mitochondrial trifunctional protein (MTF) deficiency Contact details Introduction Regional Genetics Service Long chain acyl-CoA dehydrogenase (LCHAD) deficiency / mitochondrial trifunctional Levels 4-6, Barclay House protein (MTF) deficiency is an autosomal recessive disorder of mitochondrial beta- 37 Queen Square oxidation of fatty acids. The mitochondrial trifunctional protein is composed of 4 alpha London, WC1N 3BH and 4 beta subunits, which are encoded by the HADHA and HADHB genes, respectively. It is characterized by early-onset cardiomyopathy, hypoglycemia, T +44 (0) 20 7762 6888 neuropathy, and pigmentary retinopathy, and sudden death. There is also an infantile F +44 (0) 20 7813 8578 onset form with a hepatic Reye-like syndrome, and a late-adolescent onset form with primarily a skeletal myopathy. Tandem mass spectrometry of organic acids in urine, Samples required and carnitines in blood spots, allows the diagnosis to be unequivocally determined. An 5ml venous blood in plastic EDTA additional clinical complication can occur in the pregnant mothers of affected fetuses; bottles (>1ml from neonates) they may experience maternal acute fatty liver of pregnancy (AFLP) syndrome or Prenatal testing must be arranged hypertension/haemolysis, elevated liver enzymes and low platelets (HELLP) in advance, through a Clinical syndrome. Genetics department if possible. The genes encoding the HADHA and HADHB subunits are located on chromosome Amniotic fluid or CV samples 2p23.3. The pathogenic -

CYB5R3 Gene Cytochrome B5 Reductase 3
CYB5R3 gene cytochrome b5 reductase 3 Normal Function The CYB5R3 gene provides instruction for making an enzyme called cytochrome b5 reductase 3. This enzyme is involved in transferring negatively charged particles called electrons from one molecule to another. Two versions (isoforms) of this enzyme are produced from the CYB5R3 gene. The soluble isoform is present only in red blood cells, and the membrane-bound isoform is found in all other cell types. Normal red blood cells contain molecules of iron-containing hemoglobin, which deliver oxygen to the body's tissues. The iron in hemoglobin is ferrous (Fe2+), but it can spontaneously become ferric (Fe3+). Hemoglobin that contains ferric iron is called methemoglobin, and it cannot deliver oxygen. The soluble isoform of cytochrome b5 reductase 3 changes ferric iron back to ferrous iron so hemoglobin can function. Normally, red blood cells contain less than 2 percent methemoglobin. The membrane-bound isoform is embedded in the membranes of various cellular compartments and is widely used in the body. This isoform is necessary for many chemical reactions, including the breakdown and formation of fatty acids, the formation of cholesterol, and the breakdown of various molecules and drugs. Health Conditions Related to Genetic Changes Autosomal recessive congenital methemoglobinemia More than 65 mutations in the CYB5R3 gene have been found to cause autosomal recessive congenital methemoglobinemia types I and II. Most of these CYB5R3 gene mutations cause autosomal recessive congenital methemoglobinemia type I, which is characterized by a lack of oxygen in the body's tissues and bluish appearance of the skin, lips, and nails (cyanosis). -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
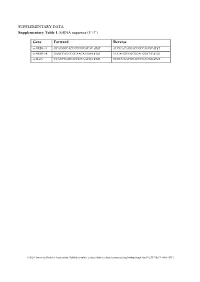
Supplementary Tables and Figures
SUPPLEMENTARY DATA Supplementary Table 1. SiRNA sequence (5’-3’) Gene Forward Reverse si-HRD1-1# GCAUGGCAGUCCUGUACAU dTdT AUGUACAGGACUGCCAUGC dTdT si-HRD1-2# GAGCCAUCCGCAACAUGAA dTdT UUCAUGUUGCGGAUGGCUC dTdT si-MafA CCAUCGAGUACGUCAACGA dTdT UCGUUGACGUACUCGAUGG dTdT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR (5’-3’) Gene Forward Reverse human HRD1 GCTCACGCCTACTACCTCAAA GCCAGACAAGTCTCTGTGACG mouse mafA AAGCGGCGCACGCTCAAGAA GGTCCCGCTCCTTGGCCAGA mouse insulin1 CACTTCCTACCCCTGCTGG ACCACAAAGATGCTGTTTGACA mouse β-actin AGGCCAACCGTGAAAAGATG AGAGCATAGCCCTCGTAGATGG human β-actin CATGTACGTTGCTATCCAGGC CTCCTTAATGTCACGCACGAT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 3. Primer sequences for ChIP (5’-3’) Gene promoter Forward Reverse mouse Insulin1, 2 GGAACTGTGAAACAGTCCAAGG CCCCCTGGACTTTGCTGTTTG ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 4. Primer sequences for PCR (5’-3’) Gene Forward Reverse HRD1-pDsred CCCAAGCTTATGTTCCGCACCGCAGT GGGGTACCCAGTGGGCAACAGGGG HRD1-pCMV- Flag GGGGTACCATGTTCCGCACCGCAGT CCCAAGCTTGTGGGCAACAGGGGACT C HRD1-pCMV-HA GGCCATGGGCCATATGGGATCCTTCC AGGGATGCCACCCGGGGATCCTCAGT GCACCGCAGTGATG GGGCAACAGGGGAC HRD1-N-HA GGCCATGGGCCATATGGGATCCTTCC -
![And Detoxification (Benzola]Pyrene Quinones/Oxygen Radicals/NADPH-Cytochrome P-450 Reductase) PAUL L](https://docslib.b-cdn.net/cover/7447/and-detoxification-benzola-pyrene-quinones-oxygen-radicals-nadph-cytochrome-p-450-reductase-paul-l-837447.webp)
And Detoxification (Benzola]Pyrene Quinones/Oxygen Radicals/NADPH-Cytochrome P-450 Reductase) PAUL L
Proc. Nati. Acad. Sci. USA Vol. 81, pp. 1696-1700, March 1984 Biochemistry Mutagenicity of quinones: Pathways of metabolic activation and detoxification (benzola]pyrene quinones/oxygen radicals/NADPH-cytochrome P-450 reductase) PAUL L. CHESIS*, DAVID E. LEVIN*, MARTYN T. SMITHt, LARS ERNSTERt, AND BRUCE N. AMES* Departments of *Biochemistry and tBiomedical and Environmental Health Sciences, School of Public Health, University of California, Berkeley, CA 94720; and tDepartment of Biochemistry, Arrhenius Laboratory, University of Stockholm, S-10691 Stockholm, Sweden Contributed by Bruce N. Ames, December 12, 1983 ABSTRACT The mutagenicity of various quinones, a class nones might also be mutagenic, and we have tested this pos- of compounds widely distributed in nature, is demonstrated in sibility using the TA104 strain, which is sensitive to a wide the Salmonella TA104 tester strain. The metabolic pathways variety of oxidative mutagens (17). We have also attempted by which four quinones, menadione, benzo[alpyrene 3,6-qui- to characterize the pathways by which several different qui- none, 9,10-phenanthrenequinone, and danthron, caused mu- nones are metabolized and to study the potential mutagenic- tagenicity in this test system were investigated in detail as were ity of the metabolites and side products formed. We there- the detoxification pathways. The two-electron reduction of fore decided to investigate only those quinones that required these quinones by NAD(P)H-quinone oxidoreductase (DT-di- metabolic activation to exhibit mutagenicity. To limit the aphorase) was not mutagenic, whereas the one-electron reduc- scope of this project we also chose not to study quinones tion, catalyzed by NADPH-cytochrome P-450 reductase, was that possess reactive leaving groups. -

Bioinformatics Analysis Based on Gene Expression Omnibus
ANTICANCER RESEARCH 39 : 1689-1698 (2019) doi:10.21873/anticanres.13274 Chemo-resistant Gastric Cancer Associated Gene Expression Signature: Bioinformatics Analysis Based on Gene Expression Omnibus JUN-BAO LIU 1* , TUNYU JIAN 2* , CHAO YUE 3, DAN CHEN 4, WEI CHEN 5, TING-TING BAO 6, HAI-XIA LIU 7, YUN CAO 8, WEI-BING LI 6, ZHIJIAN YANG 9, ROBERT M. HOFFMAN 9 and CHEN YU 6 1Traditional Chinese Medicine Department, People's Hospital of Henan Province, People's Hospital of Zhengzhou University, Zhengzhou, P.R. China; 2Institute of Botany, Jiangsu Province and Chinese Academy of Sciences, Nanjing, P.R. China; 3Department of general surgery, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, P.R. China; 4Research Center of Clinical Oncology, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, P.R. China; 5Department of Head and Neck Surgery, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, P.R. China; 6Department of Integrated TCM & Western Medicine, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, P.R. China; 7Emergency Department, The Second Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, P.R. China; 8Master candidate of Oncology, Nanjing University of Chinese Medicine, Nanjing, P.R. China; 9AntiCancer, Inc., San Diego, CA, U.S.A. Abstract. Background/Aim: This study aimed to identify identified, including 13 up-regulated and 1,473 down-regulated biomarkers for predicting the prognosis of advanced gastric genes. -

Expression in Escherichia Coli of Cytochrome C Reductase Activity from a Maize NADH:Nitrate Reductase Complementary DNA'
Plant Physiol. (1992) 99, 693-699 Received for publication September 20, 1991 0032-0889/92/99/0693/07/$01 .00/0 Accepted December 11, 1991 Expression in Escherichia coli of Cytochrome c Reductase Activity from a Maize NADH:Nitrate Reductase Complementary DNA' Wilbur H. Campbell Phytotechnology Research Center and Department of Biological Sciences, Michigan Technological University, Houghton, Michigan 49931-1295 ABSTRACT the deduced amino acid sequences of NR to related mam- A cDNA clone was isolated from a maize (Zea mays L. cv malian protein sequences because of its similarity to the Mo- W64AxW183E) scutellum Xgtl 1 library using maize leaf pterin domain of sulfite oxidase, and the C-terminal region NADH:nitrate reductase Zmnrl cDNA clone as a hybridization of NR has sequence homology to NADH:Cyt b5 reductase, probe; it was designated Zmnr1S. ZmnrlS was shown to be an which is a FAD-containing enzyme (3, 22). In between the NADH:nitrate reductase clone by nucleotide sequencing and com- Mo-pterin and FAD domains, NR has similarity to mam- parison of its deduced amino acid sequence to Zmnrl. Zmnr1S, malian Cyt b5. This central Cyt b domain of NR is bridged to which is 1.8 kilobases in length and contains the code for both the other two domains on either side of it by highly variable the cytochrome b and flavin adenine dinucleotide domains of sequence regions that appear to be hinges (3). These hinge nitrate reductase, was cloned into the EcoRI site of the Esche- regions are probably exposed in native NR and susceptible to richia coli expression vector pET5b and expressed.