Bioinformatics Analysis Based on Gene Expression Omnibus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Cytogenetic Analysis for TFE3 Rearrangement In
Modern Pathology (2014) 27, 113–127 & 2014 USCAP, Inc. All rights reserved 0893-3952/14 $32.00 113 Molecular cytogenetic analysis for TFE3 rearrangement in Xp11.2 renal cell carcinoma and alveolar soft part sarcoma: validation and clinical experience with 75 cases Jennelle C Hodge, Kathryn E Pearce, Xiaoke Wang, Anne E Wiktor, Andre M Oliveira and Patricia T Greipp Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN, USA Renal cell carcinoma with TFE3 rearrangement at Xp11.2 is a distinct subtype manifesting an indolent clinical course in children, with recent reports suggesting a more aggressive entity in adults. This subtype is morphologically heterogeneous and can be misclassified as clear cell or papillary renal cell carcinoma. TFE3 is also rearranged in alveolar soft part sarcoma. To aid in diagnosis, a break-apart strategy fluorescence in situ hybridization (FISH) probe set specific for TFE3 rearrangement and a reflex dual-color, single-fusion strategy probe set involving the most common TFE3 partner gene, ASPSCR1, were validated on formalin-fixed, paraffin- embedded tissues from nine alveolar soft part sarcoma, two suspected Xp11.2 renal cell carcinoma, and nine tumors in the differential diagnosis. The impact of tissue cut artifact was reduced through inclusion of a chromosome X centromere control probe. Analysis of the UOK-109 renal carcinoma cell line confirmed the break-apart TFE3 probe set can distinguish the subtle TFE3/NONO fusion-associated inversion of chromosome X. Subsequent extensive clinical experience was gained through analysis of 75 cases with an indication of Xp11.2 renal cell carcinoma (n ¼ 54), alveolar soft part sarcoma (n ¼ 13), perivascular epithelioid cell neoplasms (n ¼ 2), chordoma (n ¼ 1), or unspecified (n ¼ 5). -

Rap-3971 Alveolar Soft Part Sarcoma Chromosome
DATA SHEET Alveolar Soft Part Sarcoma Chromosome Region, Candidate 1 Human Item Number rAP-3971 Synonyms ASPCR1, ASPL, ASPS, RCC17, TUG, UBXD9, UBXN9, Tether containing UBX domain for GLUT4, Alveo- lar soft part sarcoma chromosomal region candidate gene 1 protein, Alveolar soft part sarcoma locus, Re- nal papillary cell carcinoma protein 17, UBX domain-containin Description ASPSCR1 Human Recombinant produced in E. coli is a single polypeptide chain containing 576 amino acids (1-553) and having a molecular mass of 62.6kDa. ASPSCR1 is fused to a 23 amino acid His-tag at N- terminus & purified by proprietary chromatographic techniques. Uniprot Accesion Number Q9BZE9 Amino Acid Sequence MGSSHHHHHH SSGLVPRGSH MGSMAAPAGG GGSAVSVLAP NGRRHTVKVT PSTVLLQVLE DTCRRQDFNP CEYDLKFQRS VLDLSLQWRF ANLPNNAKLE MVPASRSREG PENMVRIALQ LDDGS- RLQDS FCSGQTLWEL LSHFPQIREC LQHPGGATPV CVYTRDEVTG EAALRGTTLQ SLGLTGGSAT IRFVMKCYDP VGKTPGSLGS SASAGQAAAS APLPLESGEL SRGDLSRPED ADTSGPCCEH TQEKQSTRAP AAAPFVPFSG GGQRLGGPPG PTRPLTSSSA KLPKSLSSPG GPSKPKKSKS GQDPQQEQEQ ERERDPQQEQ ERERPVDREP VDREPVVCHP DLEERLQAWP AELPDEFFEL Source Escherichia Coli. Physical Appearance Sterile Filtered clear solution. Store at 4°C if entire vial will be used within 2-4 weeks. Store, frozen at -20°C and Stability for longer periods of time. For long term storage it is recommended to add a carrier protein (0.1% HSA or BSA).Avoid multiple freeze-thaw cycles. Formulation and Purity The ASPSCR1 solution (0.25mg/ml) contains 20mM Tris-HCl buffer (pH 8.0), 0.15M NaCl, 10% glycerol and 1mM DTT. Greater than 85% as determined by SDS-PAGE. Application Solubility Biological Activity Shipping Format and Condition Lyophilized powder at room temperature. Optimal dilutions should be determined by each laboratory for each application. -

CXCR4 Pathway Retards Muscle Atrophy During Cancer Cachexia
Oncogene (2016) 35, 6212–6222 © 2016 Macmillan Publishers Limited, part of Springer Nature. All rights reserved 0950-9232/16 www.nature.com/onc ORIGINAL ARTICLE Activation of the SDF1/CXCR4 pathway retards muscle atrophy during cancer cachexia GB Martinelli1, D Olivari1, AD Re Cecconi1, L Talamini1, L Ottoboni2, SH Lecker3, C Stretch4, VE Baracos4, OF Bathe5, A Resovi6, R Giavazzi1, L Cervo7 and R Piccirillo1 Cancer cachexia is a life-threatening syndrome that affects most patients with advanced cancers and causes severe body weight loss, with rapid depletion of skeletal muscle. No treatment is available. We analyzed microarray data sets to identify a subset of genes whose expression is specifically altered in cachectic muscles of Yoshida hepatoma-bearing rodents but not in those with diabetes, disuse, uremia or fasting. Ingenuity Pathways Analysis indicated that three genes belonging to the C-X-C motif chemokine receptor 4 (CXCR4) pathway were downregulated only in muscles atrophying because of cancer: stromal cell-derived factor 1 (SDF1), adenylate cyclase 7 (ADCY7), and p21 protein-activated kinase 1 (PAK1). Notably, we found that, in the Rectus Abdominis muscle of cancer patients, the expression of SDF1 and CXCR4 was inversely correlated with that of two ubiquitin ligases induced in muscle wasting, atrogin-1 and MuRF1, suggesting a possible clinical relevance of this pathway. The expression of all main SDF1 isoforms (α, β, γ) also declined in Tibialis Anterior muscle from cachectic mice bearing murine colon adenocarcinoma or human renal cancer and drugs with anticachexia properties restored their expression. Overexpressing genes of this pathway (that is, SDF1 or CXCR4) in cachectic muscles increased the fiber area by 20%, protecting them from wasting. -

CYB5R3 Gene Cytochrome B5 Reductase 3
CYB5R3 gene cytochrome b5 reductase 3 Normal Function The CYB5R3 gene provides instruction for making an enzyme called cytochrome b5 reductase 3. This enzyme is involved in transferring negatively charged particles called electrons from one molecule to another. Two versions (isoforms) of this enzyme are produced from the CYB5R3 gene. The soluble isoform is present only in red blood cells, and the membrane-bound isoform is found in all other cell types. Normal red blood cells contain molecules of iron-containing hemoglobin, which deliver oxygen to the body's tissues. The iron in hemoglobin is ferrous (Fe2+), but it can spontaneously become ferric (Fe3+). Hemoglobin that contains ferric iron is called methemoglobin, and it cannot deliver oxygen. The soluble isoform of cytochrome b5 reductase 3 changes ferric iron back to ferrous iron so hemoglobin can function. Normally, red blood cells contain less than 2 percent methemoglobin. The membrane-bound isoform is embedded in the membranes of various cellular compartments and is widely used in the body. This isoform is necessary for many chemical reactions, including the breakdown and formation of fatty acids, the formation of cholesterol, and the breakdown of various molecules and drugs. Health Conditions Related to Genetic Changes Autosomal recessive congenital methemoglobinemia More than 65 mutations in the CYB5R3 gene have been found to cause autosomal recessive congenital methemoglobinemia types I and II. Most of these CYB5R3 gene mutations cause autosomal recessive congenital methemoglobinemia type I, which is characterized by a lack of oxygen in the body's tissues and bluish appearance of the skin, lips, and nails (cyanosis). -

Genome-Wide Association and Transcriptome Studies Identify Candidate Genes and Pathways for Feed Conversion Ratio in Pigs
Miao et al. BMC Genomics (2021) 22:294 https://doi.org/10.1186/s12864-021-07570-w RESEARCH ARTICLE Open Access Genome-wide association and transcriptome studies identify candidate genes and pathways for feed conversion ratio in pigs Yuanxin Miao1,2,3, Quanshun Mei1,2, Chuanke Fu1,2, Mingxing Liao1,2,4, Yan Liu1,2, Xuewen Xu1,2, Xinyun Li1,2, Shuhong Zhao1,2 and Tao Xiang1,2* Abstract Background: The feed conversion ratio (FCR) is an important productive trait that greatly affects profits in the pig industry. Elucidating the genetic mechanisms underpinning FCR may promote more efficient improvement of FCR through artificial selection. In this study, we integrated a genome-wide association study (GWAS) with transcriptome analyses of different tissues in Yorkshire pigs (YY) with the aim of identifying key genes and signalling pathways associated with FCR. Results: A total of 61 significant single nucleotide polymorphisms (SNPs) were detected by GWAS in YY. All of these SNPs were located on porcine chromosome (SSC) 5, and the covered region was considered a quantitative trait locus (QTL) region for FCR. Some genes distributed around these significant SNPs were considered as candidates for regulating FCR, including TPH2, FAR2, IRAK3, YARS2, GRIP1, FRS2, CNOT2 and TRHDE. According to transcriptome analyses in the hypothalamus, TPH2 exhibits the potential to regulate intestinal motility through serotonergic synapse and oxytocin signalling pathways. In addition, GRIP1 may be involved in glutamatergic and GABAergic signalling pathways, which regulate FCR by affecting appetite in pigs. Moreover, GRIP1, FRS2, CNOT2,andTRHDE may regulate metabolism in various tissues through a thyroid hormone signalling pathway. -

The Orphan Disease Networks
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector ARTICLE The Orphan Disease Networks Minlu Zhang,1,3,5 Cheng Zhu,1,5 Alexis Jacomy,4 Long J. Lu,1,2,3 and Anil G. Jegga1,2,3,* The low prevalence rate of orphan diseases (OD) requires special combined efforts to improve diagnosis, prevention, and discovery of novel therapeutic strategies. To identify and investigate relationships based on shared genes or shared functional features, we have con- ducted a bioinformatic-based global analysis of all orphan diseases with known disease-causing mutant genes. Starting with a bipartite network of known OD and OD-causing mutant genes and using the human protein interactome, we first construct and topologically analyze three networks: the orphan disease network, the orphan disease-causing mutant gene network, and the orphan disease-causing mutant gene interactome. Our results demonstrate that in contrast to the common disease-causing mutant genes that are predomi- nantly nonessential, a majority of orphan disease-causing mutant genes are essential. In confirmation of this finding, we found that OD-causing mutant genes are topologically important in the protein interactome and are ubiquitously expressed. Additionally, func- tional enrichment analysis of those genes in which mutations cause ODs shows that a majority result in premature death or are lethal in the orthologous mouse gene knockout models. To address the limitations of traditional gene-based disease networks, we also construct and analyze OD networks on the basis of shared enriched features (biological processes, cellular components, pathways, phenotypes, and literature citations). -

Analysis of the Indacaterol-Regulated Transcriptome in Human Airway
Supplemental material to this article can be found at: http://jpet.aspetjournals.org/content/suppl/2018/04/13/jpet.118.249292.DC1 1521-0103/366/1/220–236$35.00 https://doi.org/10.1124/jpet.118.249292 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 366:220–236, July 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Analysis of the Indacaterol-Regulated Transcriptome in Human Airway Epithelial Cells Implicates Gene Expression Changes in the s Adverse and Therapeutic Effects of b2-Adrenoceptor Agonists Dong Yan, Omar Hamed, Taruna Joshi,1 Mahmoud M. Mostafa, Kyla C. Jamieson, Radhika Joshi, Robert Newton, and Mark A. Giembycz Departments of Physiology and Pharmacology (D.Y., O.H., T.J., K.C.J., R.J., M.A.G.) and Cell Biology and Anatomy (M.M.M., R.N.), Snyder Institute for Chronic Diseases, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada Received March 22, 2018; accepted April 11, 2018 Downloaded from ABSTRACT The contribution of gene expression changes to the adverse and activity, and positive regulation of neutrophil chemotaxis. The therapeutic effects of b2-adrenoceptor agonists in asthma was general enriched GO term extracellular space was also associ- investigated using human airway epithelial cells as a therapeu- ated with indacaterol-induced genes, and many of those, in- tically relevant target. Operational model-fitting established that cluding CRISPLD2, DMBT1, GAS1, and SOCS3, have putative jpet.aspetjournals.org the long-acting b2-adrenoceptor agonists (LABA) indacaterol, anti-inflammatory, antibacterial, and/or antiviral activity. Numer- salmeterol, formoterol, and picumeterol were full agonists on ous indacaterol-regulated genes were also induced or repressed BEAS-2B cells transfected with a cAMP-response element in BEAS-2B cells and human primary bronchial epithelial cells by reporter but differed in efficacy (indacaterol $ formoterol . -
Supplementary Tables and Figures
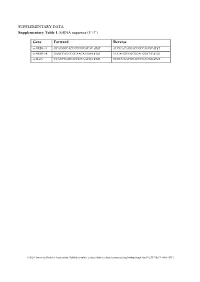
SUPPLEMENTARY DATA Supplementary Table 1. SiRNA sequence (5’-3’) Gene Forward Reverse si-HRD1-1# GCAUGGCAGUCCUGUACAU dTdT AUGUACAGGACUGCCAUGC dTdT si-HRD1-2# GAGCCAUCCGCAACAUGAA dTdT UUCAUGUUGCGGAUGGCUC dTdT si-MafA CCAUCGAGUACGUCAACGA dTdT UCGUUGACGUACUCGAUGG dTdT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR (5’-3’) Gene Forward Reverse human HRD1 GCTCACGCCTACTACCTCAAA GCCAGACAAGTCTCTGTGACG mouse mafA AAGCGGCGCACGCTCAAGAA GGTCCCGCTCCTTGGCCAGA mouse insulin1 CACTTCCTACCCCTGCTGG ACCACAAAGATGCTGTTTGACA mouse β-actin AGGCCAACCGTGAAAAGATG AGAGCATAGCCCTCGTAGATGG human β-actin CATGTACGTTGCTATCCAGGC CTCCTTAATGTCACGCACGAT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 3. Primer sequences for ChIP (5’-3’) Gene promoter Forward Reverse mouse Insulin1, 2 GGAACTGTGAAACAGTCCAAGG CCCCCTGGACTTTGCTGTTTG ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 4. Primer sequences for PCR (5’-3’) Gene Forward Reverse HRD1-pDsred CCCAAGCTTATGTTCCGCACCGCAGT GGGGTACCCAGTGGGCAACAGGGG HRD1-pCMV- Flag GGGGTACCATGTTCCGCACCGCAGT CCCAAGCTTGTGGGCAACAGGGGACT C HRD1-pCMV-HA GGCCATGGGCCATATGGGATCCTTCC AGGGATGCCACCCGGGGATCCTCAGT GCACCGCAGTGATG GGGCAACAGGGGAC HRD1-N-HA GGCCATGGGCCATATGGGATCCTTCC -

Recombinant Human ASPSCR1 Protein Catalog Number: ATGP2165
Recombinant human ASPSCR1 protein Catalog Number: ATGP2165 PRODUCT INPORMATION Expression system E.coli Domain 1-553aa UniProt No. Q9BZE9 NCBI Accession No. NP_076988 Alternative Names Tether containing uBX domain for GLuT4 isoform 1, ASPCR1, ASPL, ASPS, RCC17, TuG, uBXD9, uBXN9 PRODUCT SPECIFICATION Molecular Weight 62.6 kDa (576aa) Concentration 0.25mg/ml (determined by Bradford assay) Formulation Liquid in. 20mM Tris-HCl buffer (pH 8.0) containing 0.15M NaCl, 10% glycerol, 1mM DTT Purity > 85% by SDS-PAGE Tag His-Tag Application SDS-PAGE Storage Condition Can be stored at +2C to +8C for 1 week. For long term storage, aliquot and store at -20C to -80C. Avoid repeated freezing and thawing cycles. BACKGROUND Description ASPSCR1 contains a uBX domain and interacts with glucose transporter type 4 (GLuT4). This protein is a tether, which sequesters the GLuT4 in intracellular vesicles in muscle and fat cells in the absence of insulin, and redistributes the GLuT4 to the plasma membrane within minutes of insulin stimulation. Translocation t (X;17) (p11;q25) of this gene with transcription factor TFE3 gene results in a ASPSCR1-TFE3 fusion protein in alveolar soft part sarcoma and in renal cell carcinomas. Recombinant human ASPSCR1 protein, fused to His-tag at N- terminus, was expressed in E. coli and purified by using conventional chromatography techniques. 1 Recombinant human ASPSCR1 protein Catalog Number: ATGP2165 Amino acid Sequence MGSSHHHHHH SSGLVPRGSH MGSMAAPAGG GGSAVSVLAP NGRRHTVKVT PSTVLLQVLE DTCRRQDFNP CEYDLKFQRS VLDLSLQWRF ANLPNNAKLE -

Epigenetic Regulation of Developmental Expression of Cyp2d Genes in Mouse Liver
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Acta Pharmaceutica Sinica B 2012;2(2):146–158 Institute of Materia Medica, Chinese Academy of Medical Sciences Chinese Pharmaceutical Association Acta Pharmaceutica Sinica B www.elsevier.com/locate/apsb www.sciencedirect.com ORIGINAL ARTICLE Epigenetic regulation of developmental expression of Cyp2d genes in mouse liver Ye Lia, Xiao-bo Zhongb,n aDepartment of Pharmacology, School of Chemical Biology and Pharmaceutical Sciences, Capital Medical University, Beijing 100069, China bDepartment of Pharmacology, Toxicology, and Therapeutics, University of Kansas Medical Center, Kansas City, KS 66160, USA Received 28 November 2011; revised 26 December 2011; accepted 10 January 2012 KEY WORDS Abstract CYP2D6 expression in liver is age-dependent. Because epigenetic mechanisms, such as DNA methylation and histone modifications, modulate age-related gene expression during develop- Cyp2d; ment, and are highly conserved among species, the current study examined the epigenetic regulation of DNA methylation; age-related expression of the Cyp2d genes in mouse liver. DNA methylation (DNAme), histone 3 Histone methylation; lysine 4 dimethylation (H3K4me2), and histone 3 lysine 27 trimethylation (H3K27me3) was Liver development established by ChIP-on-chip tiling microarrays from mouse livers at prenatal, neonatal, and adult stages. Levels of DNAme, H3K4me2, and H3K27me3 were analyzed in a genomic region containing the Cyp2d clustering genes and their surrounding genes. Gradually increased expression levels of the Cyp2d9, Cyp2d10, Cyp2d22,andCyp2d26 genes from prenatal, through neonatal, to adult are associated with gradually increased levels of H3K4me2 in the nucleosomes associated with these genes. Gene expression patterns during liver development in several Cyp2d surrounding genes, such as Srebf2, Sept3, Ndufa6, Tcf2, Nfam1,andCyb5r3, could be also explained by changes of DNA methylation, H3K4me2, or H3K27me3 in those genes. -

TUG/ASPSCR1 Rabbit Pab
Leader in Biomolecular Solutions for Life Science TUG/ASPSCR1 Rabbit pAb Catalog No.: A7481 Basic Information Background Catalog No. The protein encoded by this gene contains a UBX domain and interacts with glucose A7481 transporter type 4 (GLUT4). This protein is a tether, which sequesters the GLUT4 in intracellular vesicles in muscle and fat cells in the absence of insulin, and redistributes Observed MW the GLUT4 to the plasma membrane within minutes of insulin stimulation. Translocation 80kDa t(X;17)(p11;q25) of this gene with transcription factor TFE3 gene results in a ASPSCR1- TFE3 fusion protein in alveolar soft part sarcoma and in renal cell carcinomas. Multiple Calculated MW alternatively spliced transcript variants have been found. 37kDa/54kDa/60kDa/69kDa Category Primary antibody Applications WB, IHC, IF, IP Cross-Reactivity Human, Mouse, Rat Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 79058 Q9BZE9 IHC 1:50 - 1:200 Immunogen 1:50 - 1:200 IF Recombinant fusion protein containing a sequence corresponding to amino acids 284-553 of human TUG/ASPSCR1 (NP_076988.1). IP 1:50 - 1:100 Synonyms ASPSCR1;ASPCR1;ASPL;ASPS;RCC17;TUG;UBXD9;UBXN9 Contact Product Information www.abclonal.com Source Isotype Purification Rabbit IgG Affinity purification Storage Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide,50% glycerol,pH7.3. Validation Data Western blot analysis of extracts of various cell lines, using TUG/ASPSCR1 antibody (A7481) at 1:1000 dilution. Secondary antibody: HRP Goat Anti-Rabbit IgG (H+L) (AS014) at 1:10000 dilution. Lysates/proteins: 25ug per lane. -

Epigenetic Modifications to Cytosine and Alzheimer's Disease
University of Kentucky UKnowledge Theses and Dissertations--Chemistry Chemistry 2017 EPIGENETIC MODIFICATIONS TO CYTOSINE AND ALZHEIMER’S DISEASE: A QUANTITATIVE ANALYSIS OF POST-MORTEM TISSUE Elizabeth M. Ellison University of Kentucky, [email protected] Digital Object Identifier: https://doi.org/10.13023/ETD.2017.398 Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Ellison, Elizabeth M., "EPIGENETIC MODIFICATIONS TO CYTOSINE AND ALZHEIMER’S DISEASE: A QUANTITATIVE ANALYSIS OF POST-MORTEM TISSUE" (2017). Theses and Dissertations--Chemistry. 86. https://uknowledge.uky.edu/chemistry_etds/86 This Doctoral Dissertation is brought to you for free and open access by the Chemistry at UKnowledge. It has been accepted for inclusion in Theses and Dissertations--Chemistry by an authorized administrator of UKnowledge. For more information, please contact [email protected]. STUDENT AGREEMENT: I represent that my thesis or dissertation and abstract are my original work. Proper attribution has been given to all outside sources. I understand that I am solely responsible for obtaining any needed copyright permissions. I have obtained needed written permission statement(s) from the owner(s) of each third-party copyrighted matter to be included in my work, allowing electronic distribution (if such use is not permitted by the fair use doctrine) which will be submitted to UKnowledge as Additional File. I hereby grant to The University of Kentucky and its agents the irrevocable, non-exclusive, and royalty-free license to archive and make accessible my work in whole or in part in all forms of media, now or hereafter known.