Epigenetic Regulation of Developmental Expression of Cyp2d Genes in Mouse Liver
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Retinoic Acid Induced 1 Gene Analysis in Humans and Zebrafish
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2009 RETINOIC ACID INDUCED 1 GENE ANALYSIS IN HUMANS AND ZEBRAFISH Bijal Vyas Virginia Commonwealth University Follow this and additional works at: https://scholarscompass.vcu.edu/etd Part of the Medical Genetics Commons © The Author Downloaded from https://scholarscompass.vcu.edu/etd/1901 This Thesis is brought to you for free and open access by the Graduate School at VCU Scholars Compass. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. © Bijal Akshay Vyas, 2009 All Rights Reserved RETINOIC ACID INDUCED 1 GENE ANALYSIS IN HUMANS AND ZEBRAFISH A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science at Virginia Commonwealth University. by BIJAL AKSHAY VYAS Bachelor of Biology North Carolina State University, 2007 Director: SARAH H. ELSEA Associate Professor, Department of Human and Molecular Genetics Department of Pediatrics Virginia Commonwealth University Richmond, Virginia August, 2009 Acknowledgement I would like to thank all the people who have supported and motivated me throughout my graduate studies. I had the great opportunity to work in Dr. Sarah Elsea‘s lab and I sincerely thank her for her willingness to help, patience and encouraging me after my failed experiments. I am also greatly thankful to Dr. Jim Lister for guiding and helping me every step of the way in finishing my zebrafish project. I also thank my third committee member, Dr. Andrew Davies for his project consultations, helpful alternatives and encouragement. -

Protein Kinase A-Mediated Septin7 Phosphorylation Disrupts Septin Filaments and Ciliogenesis
cells Article Protein Kinase A-Mediated Septin7 Phosphorylation Disrupts Septin Filaments and Ciliogenesis Han-Yu Wang 1,2, Chun-Hsiang Lin 1, Yi-Ru Shen 1, Ting-Yu Chen 2,3, Chia-Yih Wang 2,3,* and Pao-Lin Kuo 1,2,4,* 1 Department of Obstetrics and Gynecology, College of Medicine, National Cheng Kung University, Tainan 701, Taiwan; [email protected] (H.-Y.W.); [email protected] (C.-H.L.); [email protected] (Y.-R.S.) 2 Institute of Basic Medical Sciences, College of Medicine, National Cheng Kung University, Tainan 701, Taiwan; [email protected] 3 Department of Cell Biology and Anatomy, College of Medicine, National Cheng Kung University, Tainan 701, Taiwan 4 Department of Obstetrics and Gynecology, National Cheng-Kung University Hospital, Tainan 704, Taiwan * Correspondence: [email protected] (C.-Y.W.); [email protected] (P.-L.K.); Tel.: +886-6-2353535 (ext. 5338); (C.-Y.W.)+886-6-2353535 (ext. 5262) (P.-L.K.) Abstract: Septins are GTP-binding proteins that form heteromeric filaments for proper cell growth and migration. Among the septins, septin7 (SEPT7) is an important component of all septin filaments. Here we show that protein kinase A (PKA) phosphorylates SEPT7 at Thr197, thus disrupting septin filament dynamics and ciliogenesis. The Thr197 residue of SEPT7, a PKA phosphorylating site, was conserved among different species. Treatment with cAMP or overexpression of PKA catalytic subunit (PKACA2) induced SEPT7 phosphorylation, followed by disruption of septin filament formation. Constitutive phosphorylation of SEPT7 at Thr197 reduced SEPT7-SEPT7 interaction, but did not affect SEPT7-SEPT6-SEPT2 or SEPT4 interaction. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia. -

CYB5R3 Gene Cytochrome B5 Reductase 3
CYB5R3 gene cytochrome b5 reductase 3 Normal Function The CYB5R3 gene provides instruction for making an enzyme called cytochrome b5 reductase 3. This enzyme is involved in transferring negatively charged particles called electrons from one molecule to another. Two versions (isoforms) of this enzyme are produced from the CYB5R3 gene. The soluble isoform is present only in red blood cells, and the membrane-bound isoform is found in all other cell types. Normal red blood cells contain molecules of iron-containing hemoglobin, which deliver oxygen to the body's tissues. The iron in hemoglobin is ferrous (Fe2+), but it can spontaneously become ferric (Fe3+). Hemoglobin that contains ferric iron is called methemoglobin, and it cannot deliver oxygen. The soluble isoform of cytochrome b5 reductase 3 changes ferric iron back to ferrous iron so hemoglobin can function. Normally, red blood cells contain less than 2 percent methemoglobin. The membrane-bound isoform is embedded in the membranes of various cellular compartments and is widely used in the body. This isoform is necessary for many chemical reactions, including the breakdown and formation of fatty acids, the formation of cholesterol, and the breakdown of various molecules and drugs. Health Conditions Related to Genetic Changes Autosomal recessive congenital methemoglobinemia More than 65 mutations in the CYB5R3 gene have been found to cause autosomal recessive congenital methemoglobinemia types I and II. Most of these CYB5R3 gene mutations cause autosomal recessive congenital methemoglobinemia type I, which is characterized by a lack of oxygen in the body's tissues and bluish appearance of the skin, lips, and nails (cyanosis). -

Low Abundance of the Matrix Arm of Complex I in Mitochondria Predicts Longevity in Mice
ARTICLE Received 24 Jan 2014 | Accepted 9 Apr 2014 | Published 12 May 2014 DOI: 10.1038/ncomms4837 OPEN Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice Satomi Miwa1, Howsun Jow2, Karen Baty3, Amy Johnson1, Rafal Czapiewski1, Gabriele Saretzki1, Achim Treumann3 & Thomas von Zglinicki1 Mitochondrial function is an important determinant of the ageing process; however, the mitochondrial properties that enable longevity are not well understood. Here we show that optimal assembly of mitochondrial complex I predicts longevity in mice. Using an unbiased high-coverage high-confidence approach, we demonstrate that electron transport chain proteins, especially the matrix arm subunits of complex I, are decreased in young long-living mice, which is associated with improved complex I assembly, higher complex I-linked state 3 oxygen consumption rates and decreased superoxide production, whereas the opposite is seen in old mice. Disruption of complex I assembly reduces oxidative metabolism with concomitant increase in mitochondrial superoxide production. This is rescued by knockdown of the mitochondrial chaperone, prohibitin. Disrupted complex I assembly causes premature senescence in primary cells. We propose that lower abundance of free catalytic complex I components supports complex I assembly, efficacy of substrate utilization and minimal ROS production, enabling enhanced longevity. 1 Institute for Ageing and Health, Newcastle University, Newcastle upon Tyne NE4 5PL, UK. 2 Centre for Integrated Systems Biology of Ageing and Nutrition, Newcastle University, Newcastle upon Tyne NE4 5PL, UK. 3 Newcastle University Protein and Proteome Analysis, Devonshire Building, Devonshire Terrace, Newcastle upon Tyne NE1 7RU, UK. Correspondence and requests for materials should be addressed to T.v.Z. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Protein Interactions in the Cancer Proteome† Cite This: Mol
Molecular BioSystems View Article Online PAPER View Journal | View Issue Small-molecule binding sites to explore protein– protein interactions in the cancer proteome† Cite this: Mol. BioSyst., 2016, 12,3067 David Xu,ab Shadia I. Jalal,c George W. Sledge Jr.d and Samy O. Meroueh*aef The Cancer Genome Atlas (TCGA) offers an unprecedented opportunity to identify small-molecule binding sites on proteins with overexpressed mRNA levels that correlate with poor survival. Here, we analyze RNA-seq and clinical data for 10 tumor types to identify genes that are both overexpressed and correlate with patient survival. Protein products of these genes were scanned for binding sites that possess shape and physicochemical properties that can accommodate small-molecule probes or therapeutic agents (druggable). These binding sites were classified as enzyme active sites (ENZ), protein–protein interaction sites (PPI), or other sites whose function is unknown (OTH). Interestingly, the overwhelming majority of binding sites were classified as OTH. We find that ENZ, PPI, and OTH binding sites often occurred on the same structure suggesting that many of these OTH cavities can be used for allosteric modulation of Creative Commons Attribution 3.0 Unported Licence. enzyme activity or protein–protein interactions with small molecules. We discovered several ENZ (PYCR1, QPRT,andHSPA6)andPPI(CASC5, ZBTB32,andCSAD) binding sites on proteins that have been seldom explored in cancer. We also found proteins that have been extensively studied in cancer that have not been previously explored with small molecules that harbor ENZ (PKMYT1, STEAP3,andNNMT) and PPI (HNF4A, MEF2B,andCBX2) binding sites. All binding sites were classified by the signaling pathways to Received 29th March 2016, which the protein that harbors them belongs using KEGG. -

Characteristics of B Cell-Associated Gene Expression in Patients With
MOLECULAR MEDICINE REPORTS 13: 4113-4121, 2016 Characteristics of B cell-associated gene expression in patients with coronary artery disease WENWEN YAN*, HAOMING SONG*, JINFA JIANG, WENJUN XU, ZHU GONG, QIANGLIN DUAN, CHUANGRONG LI, YUAN XIE and LEMIN WANG Department of Internal Medicine, Division of Cardiology, Tongji Hospital, Tongji University School of Medicine, Shanghai 200065, P.R. China Received May 19, 2015; Accepted February 12, 2016 DOI: 10.3892/mmr.2016.5029 Abstract. The current study aimed to identify differentially with the two other groups. Additionally the gene expression expressed B cell-associated genes in peripheral blood mono- levels of B cell regulatory genes were measured. In patients nuclear cells and observe the changes in B cell activation at with AMI, CR1, LILRB2, LILRB3 and VAV1 mRNA expres- different stages of coronary artery disease. Groups of patients sion levels were statistically increased, whereas, CS1 and IL4I1 with acute myocardial infarction (AMI) and stable angina (SA), mRNAs were significantly reduced compared with the SA and as well as healthy volunteers, were recruited into the study control groups. There was no statistically significant difference (n=20 per group). Whole human genome microarray analysis in B cell-associated gene expression levels between patients was performed to examine the expression of B cell-associated with SA and the control group. The present study identified the genes among these three groups. The mRNA expression levels downregulation of genes associated with BCRs, B2 cells and of 60 genes associated with B cell activity and regulation were B cell regulators in patients with AMI, indicating a weakened measured using reverse transcription-quantitative polymerase T cell-B cell interaction and reduced B2 cell activation during chain reaction. -
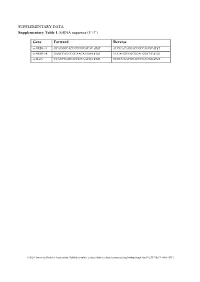
Supplementary Tables and Figures
SUPPLEMENTARY DATA Supplementary Table 1. SiRNA sequence (5’-3’) Gene Forward Reverse si-HRD1-1# GCAUGGCAGUCCUGUACAU dTdT AUGUACAGGACUGCCAUGC dTdT si-HRD1-2# GAGCCAUCCGCAACAUGAA dTdT UUCAUGUUGCGGAUGGCUC dTdT si-MafA CCAUCGAGUACGUCAACGA dTdT UCGUUGACGUACUCGAUGG dTdT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR (5’-3’) Gene Forward Reverse human HRD1 GCTCACGCCTACTACCTCAAA GCCAGACAAGTCTCTGTGACG mouse mafA AAGCGGCGCACGCTCAAGAA GGTCCCGCTCCTTGGCCAGA mouse insulin1 CACTTCCTACCCCTGCTGG ACCACAAAGATGCTGTTTGACA mouse β-actin AGGCCAACCGTGAAAAGATG AGAGCATAGCCCTCGTAGATGG human β-actin CATGTACGTTGCTATCCAGGC CTCCTTAATGTCACGCACGAT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 3. Primer sequences for ChIP (5’-3’) Gene promoter Forward Reverse mouse Insulin1, 2 GGAACTGTGAAACAGTCCAAGG CCCCCTGGACTTTGCTGTTTG ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 4. Primer sequences for PCR (5’-3’) Gene Forward Reverse HRD1-pDsred CCCAAGCTTATGTTCCGCACCGCAGT GGGGTACCCAGTGGGCAACAGGGG HRD1-pCMV- Flag GGGGTACCATGTTCCGCACCGCAGT CCCAAGCTTGTGGGCAACAGGGGACT C HRD1-pCMV-HA GGCCATGGGCCATATGGGATCCTTCC AGGGATGCCACCCGGGGATCCTCAGT GCACCGCAGTGATG GGGCAACAGGGGAC HRD1-N-HA GGCCATGGGCCATATGGGATCCTTCC -

Molecular and Epigenetic Features of Melanomas and Tumor Immune
Seremet et al. J Transl Med (2016) 14:232 DOI 10.1186/s12967-016-0990-x Journal of Translational Medicine RESEARCH Open Access Molecular and epigenetic features of melanomas and tumor immune microenvironment linked to durable remission to ipilimumab‑based immunotherapy in metastatic patients Teofila Seremet1,3*† , Alexander Koch2†, Yanina Jansen1, Max Schreuer1, Sofie Wilgenhof1, Véronique Del Marmol3, Danielle Liènard3, Kris Thielemans4, Kelly Schats5, Mark Kockx5, Wim Van Criekinge2, Pierre G. Coulie6, Tim De Meyer2, Nicolas van Baren6,7 and Bart Neyns1 Abstract Background: Ipilimumab (Ipi) improves the survival of advanced melanoma patients with an incremental long-term benefit in 10–15 % of patients. A tumor signature that correlates with this survival benefit could help optimizing indi- vidualized treatment strategies. Methods: Freshly frozen melanoma metastases were collected from patients treated with either Ipi alone (n: 7) or Ipi combined with a dendritic cell vaccine (TriMixDC-MEL) (n: 11). Samples were profiled by immunohistochemistry (IHC), whole transcriptome (RNA-seq) and methyl-DNA sequencing (MBD-seq). Results: Patients were divided in two groups according to clinical evolution: durable benefit (DB; 5 patients) and no clinical benefit (NB; 13 patients). 20 metastases were profiled by IHC and 12 were profiled by RNA- and MBD-seq. 325 genes were identified as differentially expressed between DB and NB. Many of these genes reflected a humoral and cellular immune response. MBD-seq revealed differences between DB and NB patients in the methylation of genes linked to nervous system development and neuron differentiation. DB tumors were more infiltrated by CD8+ and PD-L1+ cells than NB tumors. -

The LYR Protein Subunit NB4M/NDUFA6 of Mitochondrial Complex I Anchors an Acyl Carrier Protein and Is Essential for Catalytic Activity
The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity Heike Angerera, Michael Radermacherb, Michalina Mankowska a, Mirco Stegerc, Klaus Zwickerd, Heinrich Heidec, Ilka Wittigc,e, Ulrich Brandte,f, and Volker Zickermanna,e,1 aInstitute of Biochemistry II, Structural Bioenergetics Group, Medical School, Goethe University Frankfurt, 60438 Frankfurt, Germany; bDepartment of Molecular Physiology and Biophysics, College of Medicine, University of Vermont, Burlington, VT 05405; cFunctional Proteomics, SFB 815 Core Unit, and dInstitute of Biochemistry I, Medical School, Goethe University Frankfurt, 60590 Frankfurt, Germany; eCluster of Excellence Macromolecular Complexes, Goethe University Frankfurt, Germany; and fNijmegen Centre for Mitochondrial Disorders, Radboud University Medical Centre, 6525 GA, Nijmegen, The Netherlands Edited by Wolfgang Baumeister, Max Planck Institute of Biochemistry, Martinsried, Germany, and approved February 25, 2014 (received for review December 4, 2013) Mitochondrial complex I is the largest and most complicated enzyme is identical to LYR motif containing protein 6 (LYRM6). Sever- of the oxidative phosphorylation system. It comprises a number al other LYRM proteins were shown to be associated with of so-called accessory subunits of largely unknown structure and the maintenance of mitochondrial homeostasis. LYRM4 (alterna- function. Here we studied subunit NB4M [NDUFA6, LYR motif tive designation ISD11, iron-sulfur protein biogenesis -

The Biological Function of Human Epididymis Protein 4 in Epithelial Ovarian Cancer
University of Rhode Island DigitalCommons@URI Open Access Dissertations 2018 THE BIOLOGICAL FUNCTION OF HUMAN EPIDIDYMIS PROTEIN 4 IN EPITHELIAL OVARIAN CANCER Nicole Elizabeth James University of Rhode Island, [email protected] Follow this and additional works at: https://digitalcommons.uri.edu/oa_diss Recommended Citation James, Nicole Elizabeth, "THE BIOLOGICAL FUNCTION OF HUMAN EPIDIDYMIS PROTEIN 4 IN EPITHELIAL OVARIAN CANCER" (2018). Open Access Dissertations. Paper 753. https://digitalcommons.uri.edu/oa_diss/753 This Dissertation is brought to you for free and open access by DigitalCommons@URI. It has been accepted for inclusion in Open Access Dissertations by an authorized administrator of DigitalCommons@URI. For more information, please contact [email protected]. THE BIOLOGICAL FUNCTION OF HUMAN EPIDIDYMIS PROTEIN 4 IN EPITHELIAL OVARIAN CANCER BY NICOLE ELIZABETH JAMES A DISSERTATION SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY IN PHARMACEUTICAL SCIENCES UNIVERSITY OF RHODE ISLAND 2018 DOCTOR OF PHILOSPHY DISSERTATION OF NICOLE ELIZABETH JAMES APPROVED: Dissertation Committee: Major Professor Clinton Chichester Jennifer Ribeiro Stephen Kogut Joan Peckham Nasser H. Zawia DEAN OF THE GRADUATE SCHOOL UNIVERSITY OF RHODE ISLAND 2018 ABSTRACT Epithelial ovarian cancer (EOC) is the most common gynecologic malignancy worldwide. EOC has a notably poor prognosis, owing to the fact that patients are frequently diagnosed at a late stage after the disease has significantly progressed. While many patients typically respond well to frontline platinum-based chemotherapy, the tumor becomes chemoresistant when a recurrence follows within five years. Therefore, there is an urgent need for the discovery of non-invasive early detection biomarkers and novel targeted therapies.