RESEARCH COMMUNICATION HADHA Is a Potential Predictor Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diapositiva 1
ns 10 A Unmethylated C Methylated ) 8 Normal Normal Tissue Cervix Esophagus 4 cervical esophageal a.u. * *** * *** ( 6 TargetID MAPINFO Position Ca-Ski HeLa SW756 tissue COLO-680N KYSE-180 OACM 5.1C tissue cg04255070 22851591 TSS1500 0.896 0.070 0.082 0.060 0.613 0.622 0.075 0.020 2 (log2) 4 cg01593340 22851587 TSS1500 0.826 0.055 0.051 0.110 0.518 0.518 0.034 0.040 cg16113692 22851502 TSS200 0.964 0.073 0.072 0.030 0.625 0.785 0.018 0.010 2 0 cg26918510 22851422 TSS200 0.985 0.019 0.000 0.000 0.711 0.961 0.010 0.060 expression cg26821579 22851416 TSS200 0.988 0.083 0.058 0.000 0.387 0.955 0.021 0.050 TRMT12 mRNA Expression mRNA TRMT12 0 cg01129966 22851401 TSS200 0.933 0.039 0.077 0.020 0.815 0.963 0.078 0.010 -2 cg25421615 22851383 TSS200 0.958 0.033 0.006 0.000 0.447 0.971 0.037 0.020 mRNA cg17953636 22851321 1stExon 0.816 0.092 0.099 0.080 0.889 0.750 0.136 0.030 -4 Methylated SVIP Unmethylated SVIP relative expression (a.u.) expression relative SVIP D Cervix Esophagus HNC Meth ) HNC Unmeth Cervix Meth 3.0 *** Cervix Unmeth *** Esophagus Meth a.u. ( 2.5 CaCaEsophagus-Ski-Ski Unmeth COLO-680N Haematological Meth TSS B TSS Haematological Unmeth 2.0 -244 bp +104 bp 244 bp +104 bp 1.5 ** * expression -SVIP 1.0 HeLa KYSE-180 HeLa TSS TSS 0.5 -ACTB 244 bp +104 bp - 244 bp +104 bp mRNA 0.0 HeLa SW756 Ca-Ski SW756 SW756 OACM 5.1C TSS KYSE-180 TSS COLO-680NOACM 5.1 C - 244 bp +104 bp 244 bp +104 bp E ) 3.0 * Ca-Ski COLO-680N WT TSS TSS 2.0 a.u. -

Altered Expression and Function of Mitochondrial Я-Oxidation Enzymes
0031-3998/01/5001-0083 PEDIATRIC RESEARCH Vol. 50, No. 1, 2001 Copyright © 2001 International Pediatric Research Foundation, Inc. Printed in U.S.A. Altered Expression and Function of Mitochondrial -Oxidation Enzymes in Juvenile Intrauterine-Growth-Retarded Rat Skeletal Muscle ROBERT H. LANE, DAVID E. KELLEY, VLADIMIR H. RITOV, ANNA E. TSIRKA, AND ELISA M. GRUETZMACHER Department of Pediatrics, UCLA School of Medicine, Mattel Children’s Hospital at UCLA, Los Angeles, California 90095, U.S.A. [R.H.L.]; and Departments of Internal Medicine [D.E.K., V.H.R.] and Pediatrics [R.H.L., A.E.T., E.M.G.], University of Pittsburgh School of Medicine, Magee-Womens Research Institute, Pittsburgh, Pennsylvania 15213, U.S.A. ABSTRACT Uteroplacental insufficiency and subsequent intrauterine creased in IUGR skeletal muscle mitochondria, and isocitrate growth retardation (IUGR) affects postnatal metabolism. In ju- dehydrogenase activity was unchanged. Interestingly, skeletal venile rats, IUGR alters skeletal muscle mitochondrial gene muscle triglycerides were significantly increased in IUGR skel- expression and reduces mitochondrial NADϩ/NADH ratios, both etal muscle. We conclude that uteroplacental insufficiency alters of which affect -oxidation flux. We therefore hypothesized that IUGR skeletal muscle mitochondrial lipid metabolism, and we gene expression and function of mitochondrial -oxidation en- speculate that the changes observed in this study play a role in zymes would be altered in juvenile IUGR skeletal muscle. To test the long-term morbidity associated with IUGR. (Pediatr Res 50: this hypothesis, mRNA levels of five key mitochondrial enzymes 83–90, 2001) (carnitine palmitoyltransferase I, trifunctional protein of -oxi- dation, uncoupling protein-3, isocitrate dehydrogenase, and mi- Abbreviations tochondrial malate dehydrogenase) and intramuscular triglycer- CPTI, carnitine palmitoyltransferase I ides were quantified in 21-d-old (preweaning) IUGR and control IUGR, intrauterine growth retardation rat skeletal muscle. -

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

Role of De Novo Cholesterol Synthesis Enzymes in Cancer Jie Yang1,2, Lihua Wang1,2, Renbing Jia1,2
Journal of Cancer 2020, Vol. 11 1761 Ivyspring International Publisher Journal of Cancer 2020; 11(7): 1761-1767. doi: 10.7150/jca.38598 Review Role of de novo cholesterol synthesis enzymes in cancer Jie Yang1,2, Lihua Wang1,2, Renbing Jia1,2 1. Department of Ophthalmology, Ninth People’s Hospital of Shanghai, Shanghai Jiao Tong University School of Medicine, Shanghai, China. 2. Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai, China. Corresponding authors: Renbing Jia, [email protected] and Lihua Wang, [email protected]. Department of Ophthalmology, Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, No. 639 Zhi Zao Ju Road, Shanghai 200011, China © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See http://ivyspring.com/terms for full terms and conditions. Received: 2019.07.21; Accepted: 2019.11.30; Published: 2020.01.17 Abstract Despite extensive research in the cancer field, cancer remains one of the most prevalent diseases. There is an urgent need to identify specific targets that are safe and effective for the treatment of cancer. In recent years, cancer metabolism has come into the spotlight in cancer research. Lipid metabolism, especially cholesterol metabolism, plays a critical role in membrane synthesis as well as lipid signaling in cancer. This review focuses on the contribution of the de novo cholesterol synthesis pathway to tumorigenesis, cancer progression and metastasis. In conclusion, cholesterol metabolism could be an effective target for novel anticancer treatment. Key words: metabolic reprogramming, de novo cholesterol synthesis, cancer progress Introduction Over the past few decades, numerous published that cholesterol plays a critical role in cancer studies have focused on cancer cell metabolism and progression15-19. -

Isocitrate Dehydrogenase 1 (NADP+) (I5036)
Isocitrate Dehydrogenase 1 (NADP+), human recombinant, expressed in Escherichia coli Catalog Number I5036 Storage Temperature –20 °C CAS RN 9028-48-2 IDH1 and IDH2 have frequent genetic alterations in EC 1.1.1.42 acute myeloid leukemia4 and better understanding of Systematic name: Isocitrate:NADP+ oxidoreductase these mutations may lead to an improvement of (decarboxylating) individual cancer risk assessment.6 In addition other studies have shown loss of IDH1 in bladder cancer Synonyms: IDH1, cytosolic NADP(+)-dependent patients during tumor development suggesting this may isocitrate dehydrogenase, isocitrate:NADP+ be involved in tumor progression and metastasis.7 oxidoreductase (decarboxylating), Isocitric Dehydrogenase, ICD1, PICD, IDPC, ICDC, This product is lyophilized from a solution containing oxalosuccinate decarboxylase Tris-HCl, pH 8.0, with trehalose, ammonium sulfate, and DTT. Product Description Isocitrate dehydrogenase (NADP+) [EC 1.1.1.42] is a Purity: ³90% (SDS-PAGE) Krebs cycle enzyme, which converts isocitrate to a-ketoglutarate. The flow of isocitrate through the Specific activity: ³80 units/mg protein glyoxylate bypass is regulated by phosphorylation of isocitrate dehydrogenase, which competes for a Unit definition: 1 unit corresponds to the amount of 1 common substrate (isocitrate) with isocitrate lyase. enzyme, which converts 1 mmole of DL-isocitrate to The activity of the enzyme is dependent on the a-ketoglutarate per minute at pH 7.4 and 37 °C (NADP formation of a magnesium or manganese-isocitrate as cofactor). The activity is measured by observing the 2 complex. reduction of NADP to NADPH at 340 nm in the 7 presence of 4 mM DL-isocitrate and 2 mM MnSO4. -

Pro-Aging Effects of Xanthine Oxidoreductase Products
antioxidants Review Pro-Aging Effects of Xanthine Oxidoreductase Products , , Maria Giulia Battelli y , Massimo Bortolotti y , Andrea Bolognesi * z and Letizia Polito * z Department of Experimental, Diagnostic and Specialty Medicine-DIMES, Alma Mater Studiorum, University of Bologna, Via San Giacomo 14, 40126 Bologna, Italy; [email protected] (M.G.B.); [email protected] (M.B.) * Correspondence: [email protected] (A.B.); [email protected] (L.P.); Tel.: +39-051-20-9-4707 (A.B.); +39-051-20-9-4729 (L.P.) These authors contributed equally. y Co-last authors. z Received: 22 July 2020; Accepted: 4 September 2020; Published: 8 September 2020 Abstract: The senescence process is the result of a series of factors that start from the genetic constitution interacting with epigenetic modifications induced by endogenous and environmental causes and that lead to a progressive deterioration at the cellular and functional levels. One of the main causes of aging is oxidative stress deriving from the imbalance between the production of reactive oxygen (ROS) and nitrogen (RNS) species and their scavenging through antioxidants. Xanthine oxidoreductase (XOR) activities produce uric acid, as well as reactive oxygen and nitrogen species, which all may be relevant to such equilibrium. This review analyzes XOR activity through in vitro experiments, animal studies and clinical reports, which highlight the pro-aging effects of XOR products. However, XOR activity contributes to a regular level of ROS and RNS, which appears essential for the proper functioning of many physiological pathways. This discourages the use of therapies with XOR inhibitors, unless symptomatic hyperuricemia is present. -

(LCHAD) Deficiency / Mitochondrial Trifunctional Protein (MTF) Deficiency
Long chain acyl-CoA dehydrogenase (LCHAD) deficiency / Mitochondrial trifunctional protein (MTF) deficiency Contact details Introduction Regional Genetics Service Long chain acyl-CoA dehydrogenase (LCHAD) deficiency / mitochondrial trifunctional Levels 4-6, Barclay House protein (MTF) deficiency is an autosomal recessive disorder of mitochondrial beta- 37 Queen Square oxidation of fatty acids. The mitochondrial trifunctional protein is composed of 4 alpha London, WC1N 3BH and 4 beta subunits, which are encoded by the HADHA and HADHB genes, respectively. It is characterized by early-onset cardiomyopathy, hypoglycemia, T +44 (0) 20 7762 6888 neuropathy, and pigmentary retinopathy, and sudden death. There is also an infantile F +44 (0) 20 7813 8578 onset form with a hepatic Reye-like syndrome, and a late-adolescent onset form with primarily a skeletal myopathy. Tandem mass spectrometry of organic acids in urine, Samples required and carnitines in blood spots, allows the diagnosis to be unequivocally determined. An 5ml venous blood in plastic EDTA additional clinical complication can occur in the pregnant mothers of affected fetuses; bottles (>1ml from neonates) they may experience maternal acute fatty liver of pregnancy (AFLP) syndrome or Prenatal testing must be arranged hypertension/haemolysis, elevated liver enzymes and low platelets (HELLP) in advance, through a Clinical syndrome. Genetics department if possible. The genes encoding the HADHA and HADHB subunits are located on chromosome Amniotic fluid or CV samples 2p23.3. The pathogenic -

Lipid Metabolic Reprogramming: Role in Melanoma Progression and Therapeutic Perspectives
cancers Review Lipid metabolic Reprogramming: Role in Melanoma Progression and Therapeutic Perspectives 1, 1, 1 2 1 Laurence Pellerin y, Lorry Carrié y , Carine Dufau , Laurence Nieto , Bruno Ségui , 1,3 1, , 1, , Thierry Levade , Joëlle Riond * z and Nathalie Andrieu-Abadie * z 1 Centre de Recherches en Cancérologie de Toulouse, Equipe Labellisée Fondation ARC, Université Fédérale de Toulouse Midi-Pyrénées, Université Toulouse III Paul-Sabatier, Inserm 1037, 2 avenue Hubert Curien, tgrCS 53717, 31037 Toulouse CEDEX 1, France; [email protected] (L.P.); [email protected] (L.C.); [email protected] (C.D.); [email protected] (B.S.); [email protected] (T.L.) 2 Institut de Pharmacologie et de Biologie Structurale, CNRS, Université Toulouse III Paul-Sabatier, UMR 5089, 205 Route de Narbonne, 31400 Toulouse, France; [email protected] 3 Laboratoire de Biochimie Métabolique, CHU Toulouse, 31059 Toulouse, France * Correspondence: [email protected] (J.R.); [email protected] (N.A.-A.); Tel.: +33-582-7416-20 (J.R.) These authors contributed equally to this work. y These authors jointly supervised this work. z Received: 15 September 2020; Accepted: 23 October 2020; Published: 27 October 2020 Simple Summary: Melanoma is a devastating skin cancer characterized by an impressive metabolic plasticity. Melanoma cells are able to adapt to the tumor microenvironment by using a variety of fuels that contribute to tumor growth and progression. In this review, the authors summarize the contribution of the lipid metabolic network in melanoma plasticity and aggressiveness, with a particular attention to specific lipid classes such as glycerophospholipids, sphingolipids, sterols and eicosanoids. -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -

Glyoxysomal Malate Dehydrogenase from Watermelon Is Synthesized
Proc. Nati. Acad. Sci. USA Vol. 87, pp. 5773-5777, August 1990 Botany Glyoxysomal malate dehydrogenase from watermelon is synthesized with an amino-terminal transit peptide (isoenzymes/organelle/Citrulus vulgaris/polymerase chain reaction) CHRISTINE GIETL* Institute of Botany, Technical University of Munich, Arcisstrasse 16, D-8000 Munich 2, Federal Republic of Germany; and Department of Physiology, Carlsberg Laboratory, Gamle Carlsberg Vej 10, DK-2500 Copenhagen Valby, Denmark Communicated by Diter von Wettstein, May 11, 1990 (receivedfor review March 5, 1990) ABSTRACT The isolation and sequence of a cDNA clone and peroxisomes are seen between mitochondria and chlo- encoding the complete glyoxysomal malate dehydrogenase roplasts (2). As in plants, mammalian peroxisomes contain [gMDH; (S)-malate:NAD+ oxidoreductase, EC 1.1.1.37] of enzymes involved in the production and degradation ofH202; watermelon cotyledons are presented. Partial cDNA clones in trypanosomes, glycolysis is sequestered into microbodies were synthesized in a three part strategy, taking advantage of called glycosomes (3). All microbodies studied contain en- the polymerase chain reaction technology with oligonucleotides zymes for (3-oxidation and a specific spectrum of other based on directly determined amino acid sequences. Subse- enzymes. It is further characteristic that microbody enzyme quently, the complete done for gMDH was synthesized with a activities are also present in other cell compartments. Gly- sense primer corresponding to the nucleotide sequence of the oxysomes as well as mitochondria contain MDH, citrate N-terminal end of pre-gMDIH and an antisense primer corre- synthase, and enzymes for 83-oxidation (4). The organelle- sponding to the adenylylation site found in the mRNA. -

Cholic Acid for Treating Inborn Errors of Primary Bile Acid Synthesis NHS England Unique Reference Number URN1623 / NICE ID004
NATIONAL INSTITUTE FOR HEALTH AND CARE EXCELLENCE Clinical evidence review of cholic acid for treating inborn errors of primary bile acid synthesis NHS England unique reference number URN1623 / NICE ID004 Prepared by: NICE on behalf of NHS England Specialised Commissioning About this clinical evidence review Clinical evidence reviews are a summary of the best available evidence for a single technology within a licensed indication, for commissioning by NHS England. The clinical evidence review supports NHS England in producing clinical policies but is not NICE guidance or advice. Summary This evidence review considers cholic acid (Laboratoires CTRS [Orphacol] and Retrophin Europe Ltd [Kolbam]) for treating inborn errors of primary bile acid NICE clinical evidence review of cholic acid for treating inborn errors of primary bile acid synthesis Page 1 of 72 NHS URN1623 NICE ID004 synthesis caused by the following enzyme deficiencies in people aged 1 month and over: 3-beta-hydroxy-delta5-C27-steroid oxidoreductase (3beta-HSD) delta4-3-oxosteroid-5-beta reductase (5beta-reductase) 2- (or alpha-) methylacyl-CoA racemase (AMACR) sterol 27-hydroxylase (presenting as cerebrotendinous xanthomatosis [CTX]) cholesterol 7alpha-hydroxylase (CYP7A1). Inborn errors of primary bile acid synthesis are rare genetic conditions in which enzyme deficiencies prevent the liver from converting cholesterol in the body to bile acids (such as cholic acid and chenodeoxycholic acid). This results in the liver producing high concentrations of atypical (or ‘unusual’) bile acids and intermediary metabolites (some of which are toxic to the liver) in an attempt to establish a normal bile acid pool. Accumulation of potentially toxic atypical bile acids and metabolites, and reduced flow of bile acids may cause liver injury. -
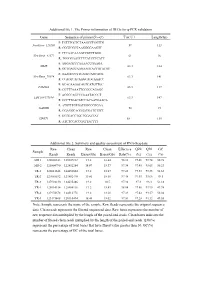
Additional File 1
Additional file 1. The Primer information of DEGs for q-PCR validation Gene Sequence of primer(5'→3') Tm(℃) Length(bp) F: TGTTTGCTCTAAGCCTGGTTG NewGene_126260 59 113 R: CGGTCGCTAAGGGGAAGTT F: CTCAACAAAGCCGTCTGGG NewGene_41572 61 96 R: TGGGGAATCTTCATCCTCATT F: AGGAGCCCAAAACCGAAGA MME 63.3 184 R: GCTGACCAAGAAGTACCGTATGT F: AAAGCCCTTCAGTCAGCACG NewGene_70974 63.3 141 R: CCAGTCACAAGCAGCAAACC F: GCACAAGGCAGTCATGTTGC FAM43A 63.3 117 R: CGTTTAAATTCCGCCAGAGC F: ACGGCAGCCCAAATACCCT LOC108177184 63.3 147 R: GCCTTGACATCCACAATGAACA F: ATGTTTGTGATGGGCGTGAA GAPDH 58 94 R: GGAGGCAGGGATGATGTTCT F: GCTGACCTGCTGGATTAT HPRT1 58 135 R: ATCTCCACCGATTACTTT Additional file 2. Summary and quality assessment of RNA-Seq data Raw Clean Raw Clean Effective Q20 Q30 GC Sample Reads Reads Bases(Gb) Bases(Gb) Rate(%) (%) (%) (%) MR-1 128008342 125607312 19.2 18.84 98.12 97.46 93.74 50.78 MR-2 125804790 122452284 18.87 18.37 97.34 97.45 93.65 50.23 TR-1 128011648 124452604 19.2 18.67 97.22 97.33 93.35 51.65 TR-2 127085532 123882470 19.06 18.58 97.48 97.45 93.66 49.5 TR-3 127994130 124635486 19.2 18.7 97.38 97.3 93.3 51.18 YR-1 128014314 125504116 19.2 18.83 98.04 97.56 93.95 49.98 YR-2 127974678 124512778 19.2 18.68 97.29 97.42 93.57 50.04 YR-3 123173408 120110494 18.48 18.02 97.51 97.28 93.32 49.55 Note: Sample represents the name of the sample. Raw Reads represents the original sequence data. Clean reads represents the filtered sequenced data. Raw bases represents the number of raw sequence data multiplied by the length of the paired-end reads.