A Phylogenomic Approach to Reconstruct Interrelationships Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi
Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 (http://www.accessscience.com/) Article by: Boschung, Herbert Department of Biological Sciences, University of Alabama, Tuscaloosa, Alabama. Gardiner, Brian Linnean Society of London, Burlington House, Piccadilly, London, United Kingdom. Publication year: 2014 DOI: http://dx.doi.org/10.1036/1097-8542.680400 (http://dx.doi.org/10.1036/1097-8542.680400) Content Morphology Euteleostei Bibliography Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi The most recent group of actinopterygians (rayfin fishes), first appearing in the Upper Triassic (Fig. 1). About 26,840 species are contained within the Teleostei, accounting for more than half of all living vertebrates and over 96% of all living fishes. Teleosts comprise 517 families, of which 69 are extinct, leaving 448 extant families; of these, about 43% have no fossil record. See also: Actinopterygii (/content/actinopterygii/009100); Osteichthyes (/content/osteichthyes/478500) Fig. 1 Cladogram showing the relationships of the extant teleosts with the other extant actinopterygians. (J. S. Nelson, Fishes of the World, 4th ed., Wiley, New York, 2006) 1 of 9 10/7/2015 1:07 PM Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 Morphology Much of the evidence for teleost monophyly (evolving from a common ancestral form) and relationships comes from the caudal skeleton and concomitant acquisition of a homocercal tail (upper and lower lobes of the caudal fin are symmetrical). This type of tail primitively results from an ontogenetic fusion of centra (bodies of vertebrae) and the possession of paired bracing bones located bilaterally along the dorsal region of the caudal skeleton, derived ontogenetically from the neural arches (uroneurals) of the ural (tail) centra. -

1 Exon Probe Sets and Bioinformatics Pipelines for All Levels of Fish Phylogenomics
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.18.949735; this version posted February 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Exon probe sets and bioinformatics pipelines for all levels of fish phylogenomics 2 3 Lily C. Hughes1,2,3,*, Guillermo Ortí1,3, Hadeel Saad1, Chenhong Li4, William T. White5, Carole 4 C. Baldwin3, Keith A. Crandall1,2, Dahiana Arcila3,6,7, and Ricardo Betancur-R.7 5 6 1 Department of Biological Sciences, George Washington University, Washington, D.C., U.S.A. 7 2 Computational Biology Institute, Milken Institute of Public Health, George Washington 8 University, Washington, D.C., U.S.A. 9 3 Department of Vertebrate Zoology, National Museum of Natural History, Smithsonian 10 Institution, Washington, D.C., U.S.A. 11 4 College of Fisheries and Life Sciences, Shanghai Ocean University, Shanghai, China 12 5 CSIRO Australian National Fish Collection, National Research Collections of Australia, 13 Hobart, TAS, Australia 14 6 Sam Noble Oklahoma Museum of Natural History, Norman, O.K., U.S.A. 15 7 Department of Biology, University of Oklahoma, Norman, O.K., U.S.A. 16 17 *Corresponding author: Lily C. Hughes, [email protected]. 18 Current address: Department of Organismal Biology and Anatomy, University of Chicago, 19 Chicago, IL. 20 21 Keywords: Actinopterygii, Protein coding, Systematics, Phylogenetics, Evolution, Target 22 capture 23 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.18.949735; this version posted February 19, 2020. -

Transcriptome Ortholog Alignment Sequence Tools (TOAST) for Phylogenomic Dataset Assembly
Transcriptome Ortholog Alignment Sequence Tools (TOAST) for Phylogenomic Dataset Assembly Dustin J. Wcisel North Carolina State University J. Thomas Howard North Carolina State University Jeffrey A. Yoder North Carolina State University Alex Dornburg ( [email protected] ) NC Museum of Natural Sciences https://orcid.org/0000-0003-0863-2283 Software Keywords: BUSCO ortholog assembly, Cetacean and teleost sh phylogeny, Missing Data Visualization, Transcriptome, Concatenated Alignment Posted Date: March 12th, 2020 DOI: https://doi.org/10.21203/rs.2.16269/v4 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at BMC Evolutionary Biology on March 30th, 2020. See the published version at https://doi.org/10.1186/s12862-020-01603-w. Page 1/18 Abstract Background Advances in next-generation sequencing technologies have reduced the cost of whole transcriptome analyses, allowing characterization of non-model species at unprecedented levels. The rapid pace of transcriptomic sequencing has driven the public accumulation of a wealth of data for phylogenomic analyses, however lack of tools aimed towards phylogeneticists to eciently identify orthologous sequences currently hinders effective harnessing of this resource. Results We introduce TOAST, an open source R software package that can utilize the ortholog searches based on the software Benchmarking Universal Single-Copy Orthologs (BUSCO) to assemble multiple sequence alignments of orthologous loci from transcriptomes for any group of organisms. By streamlining search, query, and alignment, TOAST automates the generation of locus and concatenated alignments, and also presents a series of outputs from which users can not only explore missing data patterns across their alignments, but also reassemble alignments based on user-dened acceptable missing data levels for a given research question. -

Origin, Evolution and Homologies of the Weberian Apparatus: a New Insight
Int. J. Morphol., 27(2):333-354, 2009. Origin, Evolution and Homologies of the Weberian Apparatus: A New Insight Origen, Evolución y Homologías del Aparato Weberiano: Un Nuevo Acercamiento Rui Diogo DIOGO, R. Origin, evolution and homologies of the Weberian apparatus: a new insight. Int. J. Morphol., 27(2):333-354, 2009. SUMMARY: The Weberian apparatus is essentially a mechanical device improving audition, consisting of a double chain of ossicles joining the air bladder to the inner ear. Despite being one of the most notable complex systems of teleost fishes and the subject of several comparative, developmental and functional studies, there is still much controversy concerning the origin, evolution and homologies of the structures forming this apparatus. In this paper I provide a new insight on these topics, which takes into account the results of recent works on comparative anatomy, paleontology, and ontogeny as well as of a recent extensive phylogenetic analysis including not only numerous otophysan and non-otophysan extant otocephalans but also ostariophysan fossils such as †Chanoides macropoma, †Clupavus maroccanus, †Santanichthys diasii, †Lusitanichthys characiformis, †Sorbininardus apuliensis and †Tischlingerichthys viohli. According to the evidence now available, the Weberian apparatus of otophysans seems to be the outcome of a functional integration of features acquired in basal otocephalans and in basal ostariophysans, which were very likely not directly related with the functioning of this apparatus, and of features acquired in the nodes leading to the Otophysi and to the clade including the four extant otophysan orders, which could well have been the result of a selection directly related to the functioning of the apparatus. -

Characterization of the G Protein-Coupled Receptor Family
www.nature.com/scientificreports OPEN Characterization of the G protein‑coupled receptor family SREB across fsh evolution Timothy S. Breton1*, William G. B. Sampson1, Benjamin Cliford2, Anyssa M. Phaneuf1, Ilze Smidt3, Tamera True1, Andrew R. Wilcox1, Taylor Lipscomb4,5, Casey Murray4 & Matthew A. DiMaggio4 The SREB (Super‑conserved Receptors Expressed in Brain) family of G protein‑coupled receptors is highly conserved across vertebrates and consists of three members: SREB1 (orphan receptor GPR27), SREB2 (GPR85), and SREB3 (GPR173). Ligands for these receptors are largely unknown or only recently identifed, and functions for all three are still beginning to be understood, including roles in glucose homeostasis, neurogenesis, and hypothalamic control of reproduction. In addition to the brain, all three are expressed in gonads, but relatively few studies have focused on this, especially in non‑mammalian models or in an integrated approach across the entire receptor family. The purpose of this study was to more fully characterize sreb genes in fsh, using comparative genomics and gonadal expression analyses in fve diverse ray‑fnned (Actinopterygii) species across evolution. Several unique characteristics were identifed in fsh, including: (1) a novel, fourth euteleost‑specifc gene (sreb3b or gpr173b) that likely emerged from a copy of sreb3 in a separate event after the teleost whole genome duplication, (2) sreb3a gene loss in Order Cyprinodontiformes, and (3) expression diferences between a gar species and teleosts. Overall, gonadal patterns suggested an important role for all sreb genes in teleost testicular development, while gar were characterized by greater ovarian expression that may refect similar roles to mammals. The novel sreb3b gene was also characterized by several unique features, including divergent but highly conserved amino acid positions, and elevated brain expression in pufer (Dichotomyctere nigroviridis) that more closely matched sreb2, not sreb3a. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

The Global Status of Freshwater Fish Age Validation Studies and a Prioritization Framework for Further Research Jonathan J
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Nebraska Cooperative Fish & Wildlife Research Nebraska Cooperative Fish & Wildlife Research Unit -- Staff ubP lications Unit 2015 The Global Status of Freshwater Fish Age Validation Studies and a Prioritization Framework for Further Research Jonathan J. Spurgeon University of Nebraska–Lincoln, [email protected] Martin J. Hamel University of Nebraska-Lincoln, [email protected] Kevin L. Pope U.S. Geological Survey—Nebraska Cooperative Fish and Wildlife Research Unit,, [email protected] Mark A. Pegg University of Nebraska-Lincoln, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/ncfwrustaff Part of the Aquaculture and Fisheries Commons, Environmental Indicators and Impact Assessment Commons, Environmental Monitoring Commons, Natural Resource Economics Commons, Natural Resources and Conservation Commons, and the Water Resource Management Commons Spurgeon, Jonathan J.; Hamel, Martin J.; Pope, Kevin L.; and Pegg, Mark A., "The Global Status of Freshwater Fish Age Validation Studies and a Prioritization Framework for Further Research" (2015). Nebraska Cooperative Fish & Wildlife Research Unit -- Staff Publications. 203. http://digitalcommons.unl.edu/ncfwrustaff/203 This Article is brought to you for free and open access by the Nebraska Cooperative Fish & Wildlife Research Unit at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Nebraska Cooperative Fish & Wildlife Research Unit -- Staff ubP lications by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Reviews in Fisheries Science & Aquaculture, 23:329–345, 2015 CopyrightO c Taylor & Francis Group, LLC ISSN: 2330-8249 print / 2330-8257 online DOI: 10.1080/23308249.2015.1068737 The Global Status of Freshwater Fish Age Validation Studies and a Prioritization Framework for Further Research JONATHAN J. -
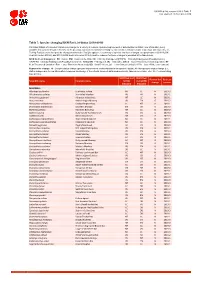
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

Percomorph Phylogeny: a Survey of Acanthomorphs and a New Proposal
BULLETIN OF MARINE SCIENCE, 52(1): 554-626, 1993 PERCOMORPH PHYLOGENY: A SURVEY OF ACANTHOMORPHS AND A NEW PROPOSAL G. David Johnson and Colin Patterson ABSTRACT The interrelationships of acanthomorph fishes are reviewed. We recognize seven mono- phyletic terminal taxa among acanthomorphs: Lampridiformes, Polymixiiformes, Paracan- thopterygii, Stephanoberyciformes, Beryciformes, Zeiformes, and a new taxon named Smeg- mamorpha. The Percomorpha, as currently constituted, are polyphyletic, and the Perciformes are probably paraphyletic. The smegmamorphs comprise five subgroups: Synbranchiformes (Synbranchoidei and Mastacembeloidei), Mugilomorpha (Mugiloidei), Elassomatidae (Elas- soma), Gasterosteiformes, and Atherinomorpha. Monophyly of Lampridiformes is justified elsewhere; we have found no new characters to substantiate the monophyly of Polymixi- iformes (which is not in doubt) or Paracanthopterygii. Stephanoberyciformes uniquely share a modification of the extrascapular, and Beryciformes a modification of the anterior part of the supraorbital and infraorbital sensory canals, here named Jakubowski's organ. Our Zei- formes excludes the Caproidae, and characters are proposed to justify the monophyly of the group in that restricted sense. The Smegmamorpha are thought to be monophyletic principally because of the configuration of the first vertebra and its intermuscular bone. Within the Smegmamorpha, the Atherinomorpha and Mugilomorpha are shown to be monophyletic elsewhere. Our Gasterosteiformes includes the syngnathoids and the Pegasiformes -

Supporting Information
Supporting Information Clarke et al. 10.1073/pnas.1607237113 Major Axes of Shape Variation and Their Anatomical a comparison: the crown teleost vs. stem teleost comparison on Correlates molecular timescales. Here, the larger SL dataset delivers a All relative warp axes that individually account for >5% of the majority of trees where crown teleosts possess significantly variation across the species sampled are displayed in Table S3. higher rates than stem teleosts, whereas the CS and pruned SL Morphospaces derived from the first three axes, containing all datasets find this result in a large minority (Fig. S3). 398 Mesozoic neopterygian species in our shape dataset, are Overall, these results suggest that choice of size metric is presented in Fig. S1. Images of sampled fossil specimens are also relatively unimportant for our dataset, and that the overall size included in Fig. S1 to illustrate the anatomical correlates of and taxonomic samplings of the dataset are more likely to in- shape axes. The positions of major neopterygian clades in mor- fluence subsequent results, despite those factors having a rela- phospace are indicated by different colors in Fig. S2. Major tively small influence here. Nevertheless, choice of metric may be teleost clades are presented in Fig. S2A and major holostean important for other datasets (e.g., different groups of organisms clades in Fig. S2B. or datasets of other biological/nonbiological structures), because RW1 captures 42.53% of the variance, reflecting changes from it is possible to envisage scenarios where the choice of size metric slender-bodied taxa to deep-bodied taxa (Fig. S1 and Table S3). -

Marine Fishes of the Azores: an Annotated Checklist and Bibliography
MARINE FISHES OF THE AZORES: AN ANNOTATED CHECKLIST AND BIBLIOGRAPHY. RICARDO SERRÃO SANTOS, FILIPE MORA PORTEIRO & JOÃO PEDRO BARREIROS SANTOS, RICARDO SERRÃO, FILIPE MORA PORTEIRO & JOÃO PEDRO BARREIROS 1997. Marine fishes of the Azores: An annotated checklist and bibliography. Arquipélago. Life and Marine Sciences Supplement 1: xxiii + 242pp. Ponta Delgada. ISSN 0873-4704. ISBN 972-9340-92-7. A list of the marine fishes of the Azores is presented. The list is based on a review of the literature combined with an examination of selected specimens available from collections of Azorean fishes deposited in museums, including the collection of fish at the Department of Oceanography and Fisheries of the University of the Azores (Horta). Personal information collected over several years is also incorporated. The geographic area considered is the Economic Exclusive Zone of the Azores. The list is organised in Classes, Orders and Families according to Nelson (1994). The scientific names are, for the most part, those used in Fishes of the North-eastern Atlantic and the Mediterranean (FNAM) (Whitehead et al. 1989), and they are organised in alphabetical order within the families. Clofnam numbers (see Hureau & Monod 1979) are included for reference. Information is given if the species is not cited for the Azores in FNAM. Whenever available, vernacular names are presented, both in Portuguese (Azorean names) and in English. Synonyms, misspellings and misidentifications found in the literature in reference to the occurrence of species in the Azores are also quoted. The 460 species listed, belong to 142 families; 12 species are cited for the first time for the Azores. -

Systematic Morphology of Fishes in the Early 21St Century
Copeia 103, No. 4, 2015, 858–873 When Tradition Meets Technology: Systematic Morphology of Fishes in the Early 21st Century Eric J. Hilton1, Nalani K. Schnell2, and Peter Konstantinidis1 Many of the primary groups of fishes currently recognized have been established through an iterative process of anatomical study and comparison of fishes that has spanned a time period approaching 500 years. In this paper we give a brief history of the systematic morphology of fishes, focusing on some of the individuals and their works from which we derive our own inspiration. We further discuss what is possible at this point in history in the anatomical study of fishes and speculate on the future of morphology used in the systematics of fishes. Beyond the collection of facts about the anatomy of fishes, morphology remains extremely relevant in the age of molecular data for at least three broad reasons: 1) new techniques for the preparation of specimens allow new data sources to be broadly compared; 2) past morphological analyses, as well as new ideas about interrelationships of fishes (based on both morphological and molecular data) provide rich sources of hypotheses to test with new morphological investigations; and 3) the use of morphological data is not limited to understanding phylogeny and evolution of fishes, but rather is of broad utility to understanding the general biology (including phenotypic adaptation, evolution, ecology, and conservation biology) of fishes. Although in some ways morphology struggles to compete with the lure of molecular data for systematic research, we see the anatomical study of fishes entering into a new and exciting phase of its history because of recent technological and methodological innovations.