Fish-T1K (Transcriptomes of 1,000 Fishes) Project: Large-Scale Transcriptome Data for Fish Evolution Studies Ying Sun1,2*†, Yu Huang2†, Xiaofeng Li2†, Carole C
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Article Evolutionary Dynamics of the OR Gene Repertoire in Teleost Fishes
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.09.434524; this version posted March 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Article Evolutionary dynamics of the OR gene repertoire in teleost fishes: evidence of an association with changes in olfactory epithelium shape Maxime Policarpo1, Katherine E Bemis2, James C Tyler3, Cushla J Metcalfe4, Patrick Laurenti5, Jean-Christophe Sandoz1, Sylvie Rétaux6 and Didier Casane*,1,7 1 Université Paris-Saclay, CNRS, IRD, UMR Évolution, Génomes, Comportement et Écologie, 91198, Gif-sur-Yvette, France. 2 NOAA National Systematics Laboratory, National Museum of Natural History, Smithsonian Institution, Washington, D.C. 20560, U.S.A. 3Department of Paleobiology, National Museum of Natural History, Smithsonian Institution, Washington, D.C., 20560, U.S.A. 4 Independent Researcher, PO Box 21, Nambour QLD 4560, Australia. 5 Université de Paris, Laboratoire Interdisciplinaire des Energies de Demain, Paris, France 6 Université Paris-Saclay, CNRS, Institut des Neurosciences Paris-Saclay, 91190, Gif-sur- Yvette, France. 7 Université de Paris, UFR Sciences du Vivant, F-75013 Paris, France. * Corresponding author: e-mail: [email protected]. !1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.09.434524; this version posted March 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Teleost fishes perceive their environment through a range of sensory modalities, among which olfaction often plays an important role. -

Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi
Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 (http://www.accessscience.com/) Article by: Boschung, Herbert Department of Biological Sciences, University of Alabama, Tuscaloosa, Alabama. Gardiner, Brian Linnean Society of London, Burlington House, Piccadilly, London, United Kingdom. Publication year: 2014 DOI: http://dx.doi.org/10.1036/1097-8542.680400 (http://dx.doi.org/10.1036/1097-8542.680400) Content Morphology Euteleostei Bibliography Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi The most recent group of actinopterygians (rayfin fishes), first appearing in the Upper Triassic (Fig. 1). About 26,840 species are contained within the Teleostei, accounting for more than half of all living vertebrates and over 96% of all living fishes. Teleosts comprise 517 families, of which 69 are extinct, leaving 448 extant families; of these, about 43% have no fossil record. See also: Actinopterygii (/content/actinopterygii/009100); Osteichthyes (/content/osteichthyes/478500) Fig. 1 Cladogram showing the relationships of the extant teleosts with the other extant actinopterygians. (J. S. Nelson, Fishes of the World, 4th ed., Wiley, New York, 2006) 1 of 9 10/7/2015 1:07 PM Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 Morphology Much of the evidence for teleost monophyly (evolving from a common ancestral form) and relationships comes from the caudal skeleton and concomitant acquisition of a homocercal tail (upper and lower lobes of the caudal fin are symmetrical). This type of tail primitively results from an ontogenetic fusion of centra (bodies of vertebrae) and the possession of paired bracing bones located bilaterally along the dorsal region of the caudal skeleton, derived ontogenetically from the neural arches (uroneurals) of the ural (tail) centra. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

The Divergent Genomes of Teleosts
Postprint copy Annu. Rev. Anim. Biosci. 2018. 6:X--X https://doi.org/10.1146/annurev-animal-030117-014821 Copyright © 2018 by Annual Reviews. All rights reserved RAVI ■ VENKATESH DIVERGENT GENOMES OF TELEOSTS THE DIVERGENT GENOMES OF TELEOSTS Vydianathan Ravi and Byrappa Venkatesh Institute of Molecular and Cell Biology, A*STAR (Agency for Science, Technology and Research), Biopolis, Singapore 138673, Singapore; email: [email protected], [email protected] ■ Abstract Boasting nearly 30,000 species, teleosts account for half of all living vertebrates and approximately 98% of all ray-finned fish species (Actinopterygii). Teleosts are also the largest and most diverse group of vertebrates, exhibiting an astonishing level of morphological, physiological, and behavioral diversity. Previous studies had indicated that the teleost lineage has experienced an additional whole-genome duplication event. Recent comparative genomic analyses of teleosts and other bony vertebrates using spotted gar (a nonteleost ray-finned fish) and elephant shark (a cartilaginous fish) as outgroups have revealed several divergent features of teleost genomes. These include an accelerated evolutionary rate of protein-coding and nucleotide sequences, a higher rate of intron turnover, and loss of many potential cis-regulatory elements and shorter conserved syntenic blocks. A combination of these divergent genomic features might have contributed to the evolution of the amazing phenotypic diversity and morphological innovations of teleosts. Keywords whole-genome duplication, evolutionary rate, intron turnover, conserved noncoding elements, conserved syntenic blocks, phenotypic diversity INTRODUCTION With over 68,000 known species (IUCN 2017; http://www.iucnredlist.org), vertebrates are the most dominant and successful group of animals on earth, inhabiting both terrestrial and aquatic habitats. -

Betanodavirus and VER Disease: a 30-Year Research Review
pathogens Review Betanodavirus and VER Disease: A 30-year Research Review Isabel Bandín * and Sandra Souto Departamento de Microbioloxía e Parasitoloxía-Instituto de Acuicultura, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain; [email protected] * Correspondence: [email protected] Received: 20 December 2019; Accepted: 4 February 2020; Published: 9 February 2020 Abstract: The outbreaks of viral encephalopathy and retinopathy (VER), caused by nervous necrosis virus (NNV), represent one of the main infectious threats for marine aquaculture worldwide. Since the first description of the disease at the end of the 1980s, a considerable amount of research has gone into understanding the mechanisms involved in fish infection, developing reliable diagnostic methods, and control measures, and several comprehensive reviews have been published to date. This review focuses on host–virus interaction and epidemiological aspects, comprising viral distribution and transmission as well as the continuously increasing host range (177 susceptible marine species and epizootic outbreaks reported in 62 of them), with special emphasis on genotypes and the effect of global warming on NNV infection, but also including the latest findings in the NNV life cycle and virulence as well as diagnostic methods and VER disease control. Keywords: nervous necrosis virus (NNV); viral encephalopathy and retinopathy (VER); virus–host interaction; epizootiology; diagnostics; control 1. Introduction Nervous necrosis virus (NNV) is the causative agent of viral encephalopathy and retinopathy (VER), otherwise known as viral nervous necrosis (VNN). The disease was first described at the end of the 1980s in Australia and in the Caribbean [1–3], and has since caused a great deal of mortalities and serious economic losses in a variety of reared marine fish species, but also in freshwater species worldwide. -

Concentración Y Tiempo Máximo De Exposición De Juveniles De Pargo
State of research of the Osteichthyes fish related to coral reefs in the Honduran Caribbean with catalogued records Estado del conocimiento de los peces osteíctios asociados a los arrecifes de coral en el Caribe de Honduras, con registros catalogados Anarda Isabel Salgado Ordoñez1, Julio Enrique Mérida Colindres1* & Gustavo Adolfo Cruz1 ABSTRACT Research on Honduran coral reef fish has been isolated and scattered. A list of fish species related to coral reefs was consolidated to establish a compiled database with updated taxonomy. The study was conducted between October 2017 and December 2018. Using primary and secondary sources, all potential species in the Western Atlantic were considered, and their actual presence was confirmed using catalogued records published in peer-reviewed journals that included Honduras. In addition, the specimens kept in the Museum of Natural History of Universidad Nacional Autónoma de Honduras were added. Once the list was consolidated, the taxonomic status of each species was updated based on recent literature. A total of 159 species and 76 genera were registered in 32 families. The family with the most species was Labrisomidae with 27 species (17%). Five families had more than five 5 genera registered, while four 4 were represented by more than 16 species, which is equivalent to 42% genera and 51% species. Gobiidae was represented by 10 genera (13%) and 21 species (13%), of which two 2 were endemic: Tigrigobius rubrigenis and Elacatinus lobeli. In turn, Grammatidae was represented by one endemic species Lipogramma idabeli (1.8%). The species Diodon holocanthus and Sphoeroides testudineus represent the first catalogued records for Honduras. -

Euteleostei: Aulopiformes) and the Timing of Deep-Sea Adaptations ⇑ Matthew P
Molecular Phylogenetics and Evolution 57 (2010) 1194–1208 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Estimating divergence times of lizardfishes and their allies (Euteleostei: Aulopiformes) and the timing of deep-sea adaptations ⇑ Matthew P. Davis a, , Christopher Fielitz b a Museum of Natural Science, Louisiana State University, 119 Foster Hall, Baton Rouge, LA 70803, USA b Department of Biology, Emory & Henry College, Emory, VA 24327, USA article info abstract Article history: The divergence times of lizardfishes (Euteleostei: Aulopiformes) are estimated utilizing a Bayesian Received 18 May 2010 approach in combination with knowledge of the fossil record of teleosts and a taxonomic review of fossil Revised 1 September 2010 aulopiform taxa. These results are integrated with a study of character evolution regarding deep-sea evo- Accepted 7 September 2010 lutionary adaptations in the clade, including simultaneous hermaphroditism and tubular eyes. Diver- Available online 18 September 2010 gence time estimations recover that the stem species of the lizardfishes arose during the Early Cretaceous/Late Jurassic in a marine environment with separate sexes, and laterally directed, round eyes. Keywords: Tubular eyes have arisen independently at different times in three deep-sea pelagic predatory aulopiform Phylogenetics lineages. Simultaneous hermaphroditism evolved a single time in the stem species of the suborder Character evolution Deep-sea Alepisauroidei, the clade of deep-sea aulopiforms during the Early Cretaceous. This result indicates the Euteleostei oldest known evolutionary event of simultaneous hermaphroditism in vertebrates, with the Alepisauroidei Hermaphroditism being the largest vertebrate clade with this reproductive strategy. Divergence times Ó 2010 Elsevier Inc. -

DEEPFISHMAN Document 5 : Review of Parasites, Pathogens
DEEPFISHMAN Management And Monitoring Of Deep-sea Fisheries And Stocks Project number: 227390 Small or medium scale focused research action Topic: FP7-KBBE-2008-1-4-02 (Deepsea fisheries management) DEEPFISHMAN Document 5 Title: Review of parasites, pathogens and contaminants of deep sea fish with a focus on their role in population health and structure Due date: none Actual submission date: 10 June 2010 Start date of the project: April 1st, 2009 Duration : 36 months Organization Name of lead coordinator: Ifremer Dissemination Level: PU (Public) Date: 10 June 2010 Review of parasites, pathogens and contaminants of deep sea fish with a focus on their role in population health and structure. Matt Longshaw & Stephen Feist Cefas Weymouth Laboratory Barrack Road, The Nothe, Weymouth, Dorset DT4 8UB 1. Introduction This review provides a summary of the parasites, pathogens and contaminant related impacts on deep sea fish normally found at depths greater than about 200m There is a clear focus on worldwide commercial species but has an emphasis on records and reports from the north east Atlantic. In particular, the focus of species following discussion were as follows: deep-water squalid sharks (e.g. Centrophorus squamosus and Centroscymnus coelolepis), black scabbardfish (Aphanopus carbo) (except in ICES area IX – fielded by Portuguese), roundnose grenadier (Coryphaenoides rupestris), orange roughy (Hoplostethus atlanticus), blue ling (Molva dypterygia), torsk (Brosme brosme), greater silver smelt (Argentina silus), Greenland halibut (Reinhardtius hippoglossoides), deep-sea redfish (Sebastes mentella), alfonsino (Beryx spp.), red blackspot seabream (Pagellus bogaraveo). However, it should be noted that in some cases no disease or contaminant data exists for these species. -

Fishes-Of-The-Salish-Sea-Pp18.Pdf
NOAA Professional Paper NMFS 18 Fishes of the Salish Sea: a compilation and distributional analysis Theodore W. Pietsch James W. Orr September 2015 U.S. Department of Commerce NOAA Professional Penny Pritzker Secretary of Commerce Papers NMFS National Oceanic and Atmospheric Administration Kathryn D. Sullivan Scientifi c Editor Administrator Richard Langton National Marine Fisheries Service National Marine Northeast Fisheries Science Center Fisheries Service Maine Field Station Eileen Sobeck 17 Godfrey Drive, Suite 1 Assistant Administrator Orono, Maine 04473 for Fisheries Associate Editor Kathryn Dennis National Marine Fisheries Service Offi ce of Science and Technology Fisheries Research and Monitoring Division 1845 Wasp Blvd., Bldg. 178 Honolulu, Hawaii 96818 Managing Editor Shelley Arenas National Marine Fisheries Service Scientifi c Publications Offi ce 7600 Sand Point Way NE Seattle, Washington 98115 Editorial Committee Ann C. Matarese National Marine Fisheries Service James W. Orr National Marine Fisheries Service - The NOAA Professional Paper NMFS (ISSN 1931-4590) series is published by the Scientifi c Publications Offi ce, National Marine Fisheries Service, The NOAA Professional Paper NMFS series carries peer-reviewed, lengthy original NOAA, 7600 Sand Point Way NE, research reports, taxonomic keys, species synopses, fl ora and fauna studies, and data- Seattle, WA 98115. intensive reports on investigations in fi shery science, engineering, and economics. The Secretary of Commerce has Copies of the NOAA Professional Paper NMFS series are available free in limited determined that the publication of numbers to government agencies, both federal and state. They are also available in this series is necessary in the transac- exchange for other scientifi c and technical publications in the marine sciences. -

The Exceptional Diversity of Visual Adaptations in Deep-Sea Teleost Fishes
Seminars in Cell and Developmental Biology xxx (xxxx) xxx–xxx Contents lists available at ScienceDirect Seminars in Cell & Developmental Biology journal homepage: www.elsevier.com/locate/semcdb Review The exceptional diversity of visual adaptations in deep-sea teleost fishes Fanny de Busserolles*, Lily Fogg, Fabio Cortesi, Justin Marshall Queensland Brain Institute, The University of Queensland, St Lucia, Queensland 4072, Australia ARTICLE INFO ABSTRACT Keywords: The deep-sea is the largest and one of the dimmest habitats on earth. In this extreme environment, every photon Deep-sea teleost counts and may make the difference between life and death for its inhabitants. Two sources of light are present Dim-light vision in the deep-sea; downwelling light, that becomes dimmer and spectrally narrower with increasing depth until Ocular adaptation completely disappearing at around 1000 m, and bioluminescence, the light emitted by animals themselves. Retina Despite these relatively dark and inhospitable conditions, many teleost fish have made the deep-sea their home, Opsin relying heavily on vision to survive. Their visual systems have had to adapt, sometimes in astonishing and Bioluminescence bizarre ways. This review examines some aspects of the visual system of deep-sea teleosts and highlights the exceptional diversity in both optical and retinal specialisations. We also reveal how widespread several of these adaptations are across the deep-sea teleost phylogeny. Finally, the significance of some recent findings as well as the surprising diversity in visual adaptations is discussed. 1. Introduction or mate detection, to communicate, camouflage, or for navigation, in- cluding to stay within a particular depth range [4,5]. -
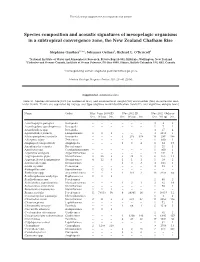
Marine Ecology Progress Series 503:23
The following supplement accompanies the article 1 Species composition and acoustic signatures of mesopelagic organisms in a subtropical convergence zone, the New Zealand Chatham Rise Stéphane Gauthier1, 2,*, Johannes Oeffner1, Richard L. O’Driscoll1 1National Institute of Water and Atmospheric Research, Private Bag 14-901, Kilbirnie, Wellington, New Zealand 2Fisheries and Oceans Canada, Institute of Ocean Sciences, PO Box 6000, Sidney, British Columbia V8L 4B2, Canada *Corresponding author: [email protected] Marine Ecology Progress Series: 503: 23–40 (2014) Supplement. Additional data Table S1. Species occurrence (Occ.) in number of tows, and abundance in weight (Wt) and number (No.) in successful mid - water trawls. Trawls are separated by voyage and type (daytime mark identification trawls ID, and nighttime oblique tows) Name Order May−June 2008 ID Nov 2011 ID Nov 2011 Oblique Occ. Wt (g) No. Occ. Wt (g) No. Occ. Wt (g) No. Acanthephyra pelagica Decapoda − − − − − − 3 9 3 Acanthephyra quadrispinosa Decapoda − − − − − − 1 7 1 Acanthephyra spp. Decapoda − − − − − − 3 17 4 Agrostichthys parkeri Lampriformes 1 8 1 − − − 3 1813 3 Alainopasiphaea australis Decapoda − − − 3 273 356 14 287 366 Allocyttus niger Zeiformes − − − − − − 1 600 1 Amphipod (unspecified) Amphipoda − − − 1 3 4 8 34 17 Anoplogaster cornuta Beryciformes − − − − − − 1 25 1 Apristurus spp. Carcharhiniformes − − − − − − 1 900 1 Argentina elongata Argentiniformes − − − − − − 4 541 6 Argyropelecus gigas Stomiiformes 2 20 2 2 38 5 8 165 12 Argyropelecus hemigymnus Stomiiformes 4 12 6 1 1 1 1 19 2 Astronesthes spp. Stomiiformes − − − 1 3 3 3 103 3 Atolla wyvillei Coronatae − − − − − − 4 83 7 Bathophilus ater Stomiiformes 1 12 1 − − − − − − Bathylagus spp. -

A Rare Occurrence of Matched Otoliths And
A RARE OCCURRENCE OF MATCHED OTOLITHS AND ASSOCIATED SKELETAL REMAINS OF APOGON TOWNSENDI (OSTEICHTHYES) FROM THE CALOOSAHATCHEE FORMATION (LOWER PLEISTOCENE) OF FLORIDA Gary L. Stringer1, Richard C. Hulbert Jr.2, Dirk Nolf3, Paul Roth4, and Roger W. Portell4 ABSTRACT A matched pair of otoliths (right and left saccular otoliths) and associated skeletal remains (n = 107) of Apogon townsendi (belted cardinalfish) were obtained in unconsolidated sediment from inside the valves of an articulated scallop Carolinapecten eboreus. The scallop specimen was collected in Hendry County, Florida, from the lower Pleistocene Caloosahatchee Formation, approximately 1.7 to 2.1 Ma. The recov- ery of this vertebrate material is highly significant because no skeletal remains of bony fish with in situ or associated otoliths are known from the Gulf or Atlantic coasts of the United States. Furthermore, the specimen represents the first fossil record of the family Apogonidae and the genus Apogon from Florida and the first report of the species Apogon townsendi in the fossil record. The length of the fossil Apogon townsendi was determined to be 4.7 cm based on the linear relationship between fish length and otolith length and utilizing modern specimens of the species for comparison and analysis. The length of the fossil Apogon townsendi indicated that it was an adult fish upon death (> 2.1 cm). Although several taphonomic scenarios are considered, including commensalism, it is believed that the apogonid died in close proximity to the empty scallop shell, which was followed by fairly rapid washing in of sediment with the fish into the valves of the scallop (i.e., sediment trapping).