Betanodavirus and VER Disease: a 30-Year Research Review
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Differential Segregation of Nodaviral Coat Protein and RNA Into Progeny Virions During Mixed Infection with FHV and Nov
Virology 454-455 (2014) 280–290 Contents lists available at ScienceDirect Virology journal homepage: www.elsevier.com/locate/yviro Differential segregation of nodaviral coat protein and RNA into progeny virions during mixed infection with FHV and NoV Radhika Gopal, P. Arno Venter 1, Anette Schneemann n Department of Cell and Molecular Biology, The Scripps Research Institute, La Jolla, CA, USA article info abstract Article history: Nodaviruses are icosahedral viruses with a bipartite, positive-sense RNA genome. The two RNAs are Received 30 December 2013 packaged into a single virion by a poorly understood mechanism. We chose two distantly related Returned to author for revisions nodaviruses, Flock House virus and Nodamura virus, to explore formation of viral reassortants as a 27 January 2014 means to further understand genome recognition and encapsidation. In mixed infections, the viruses Accepted 3 March 2014 were incompatible at the level of RNA replication and their coat proteins segregated into separate Available online 21 March 2014 populations of progeny particles. RNA packaging, on the other hand, was indiscriminate as all four viral Keywords: RNAs were detectable in each progeny population. Consistent with the trans-encapsidation phenotype, Flock House virus fluorescence in situ hybridization of viral RNA revealed that the genomes of the two viruses co-localized Nodamura virus throughout the cytoplasm. Our results imply that nodaviral RNAs lack rigorously defined packaging Mixed infection signals and that co-encapsidation of the viral RNAs does not require a pair of cognate RNA1 and RNA2. Viral assembly & RNA encapsidation 2014 Elsevier Inc. All rights reserved. Viral reassortant Introduction invaginations of the outer membrane of the organelle (Kopek et al., 2007). -

Comprehensive Phylogeny of Ray-Finned Fishes (Actinopterygii) Based on Transcriptomic and Genomic Data
Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data Lily C. Hughesa,b,1,2, Guillermo Ortía,b,1,2, Yu Huangc,d,1, Ying Sunc,e,1, Carole C. Baldwinb, Andrew W. Thompsona,b, Dahiana Arcilaa,b, Ricardo Betancur-R.b,f, Chenhong Lig, Leandro Beckerh, Nicolás Bellorah, Xiaomeng Zhaoc,d, Xiaofeng Lic,d, Min Wangc, Chao Fangd, Bing Xiec, Zhuocheng Zhoui, Hai Huangj, Songlin Chenk, Byrappa Venkateshl,2, and Qiong Shic,d,2 aDepartment of Biological Sciences, The George Washington University, Washington, DC 20052; bNational Museum of Natural History, Smithsonian Institution, Washington, DC 20560; cShenzhen Key Lab of Marine Genomics, Guangdong Provincial Key Lab of Molecular Breeding in Marine Economic Animals, Beijing Genomics Institute Academy of Marine Sciences, Beijing Genomics Institute Marine, Beijing Genomics Institute, 518083 Shenzhen, China; dBeijing Genomics Institute Education Center, University of Chinese Academy of Sciences, 518083 Shenzhen, China; eChina National GeneBank, Beijing Genomics Institute-Shenzhen, 518120 Shenzhen, China; fDepartment of Biology, University of Puerto Rico–Rio Piedras, San Juan 00931, Puerto Rico; gKey Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Shanghai Ocean University, Ministry of Education, 201306 Shanghai, China; hLaboratorio de Ictiología y Acuicultura Experimental, Universidad Nacional del Comahue–CONICET, 8400 Bariloche, Argentina; iProfessional Committee of Native Aquatic Organisms and Water Ecosystem, China Fisheries Association, 100125 Beijing, China; jCollege of Life Science and Ecology, Hainan Tropical Ocean University, 572022 Sanya, China; kYellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, 266071 Qingdao, China; and lComparative Genomics Laboratory, Institute of Molecular and Cell Biology, A*STAR, Biopolis, 138673 Singapore Edited by Scott V. -

Article Evolutionary Dynamics of the OR Gene Repertoire in Teleost Fishes
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.09.434524; this version posted March 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Article Evolutionary dynamics of the OR gene repertoire in teleost fishes: evidence of an association with changes in olfactory epithelium shape Maxime Policarpo1, Katherine E Bemis2, James C Tyler3, Cushla J Metcalfe4, Patrick Laurenti5, Jean-Christophe Sandoz1, Sylvie Rétaux6 and Didier Casane*,1,7 1 Université Paris-Saclay, CNRS, IRD, UMR Évolution, Génomes, Comportement et Écologie, 91198, Gif-sur-Yvette, France. 2 NOAA National Systematics Laboratory, National Museum of Natural History, Smithsonian Institution, Washington, D.C. 20560, U.S.A. 3Department of Paleobiology, National Museum of Natural History, Smithsonian Institution, Washington, D.C., 20560, U.S.A. 4 Independent Researcher, PO Box 21, Nambour QLD 4560, Australia. 5 Université de Paris, Laboratoire Interdisciplinaire des Energies de Demain, Paris, France 6 Université Paris-Saclay, CNRS, Institut des Neurosciences Paris-Saclay, 91190, Gif-sur- Yvette, France. 7 Université de Paris, UFR Sciences du Vivant, F-75013 Paris, France. * Corresponding author: e-mail: [email protected]. !1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.09.434524; this version posted March 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Teleost fishes perceive their environment through a range of sensory modalities, among which olfaction often plays an important role. -

ABSTRACT LUCKENBACH, JOHN ADAM. Breeding Biotechnology
ABSTRACT LUCKENBACH, JOHN ADAM. Breeding Biotechnology, Sex Determination, and Growth in Southern flounder, Paralichthys lethostigma. (Under the direction of Dr. John Godwin and Dr. Russell J. Borski). Southern flounder (Paralichthys lethostigma) support valuable, but declining US fisheries. This species is therefore a strong candidate for aquaculture to mitigate fishing impacts and stabilize seafood supply. Because female flounder reach substantially larger sizes than males, all-female culture is desirable for commercial aquaculture. Hence, a thorough understanding of sexual development, its timing and regulation by temperature is essential for optimization of flounder aquaculture. To better understand ovarian and testicular development in southern flounder, structural and cellular sex-distinguishing markers were studied using histological methods. We found that histologically discernible sex differentiation occurs in southern flounder at ~75-120 mm TL and that early differentiation is considerably delayed relative to its Japanese congener, P. olivaceus. High (28°C) and low (18°C) water temperatures, produced a higher proportion of males (96% and 78% males, respectively). The sex ratio at a mid-range (23°C) temperature was not different from 1:1. This suggests that southern flounder possess a temperature sensitive mechanism of sex determination. Growth was also affected by temperature with the temperature that maximized females inducing better growth. Aromatase cytochrome P450 (P450arom) is responsible for estrogen biosynthesis and plays a critical role in ovarian differentiation. We cloned ovarian P450arom and developed a qRT-PCR for assessment of early sex differentiation. The deduced amino acid sequence for southern flounder P450arom is very similar to P450arom in other teleosts. Comparison of P450arom intron sequences of southern flounder within and between different populations revealed substantial inter-individual variation that may affect sex determination responses. -

Atheriniformes : Atherinidae
Atheriniformes: Atherinidae 2111 Atheriniformes: Atherinidae Order ATHERINIFORMES ATHERINIDAE Silversides by L. Tito de Morais, IRD/LEMAR, University of Brest, Plouzané, France; M. Sylla, Centre de Recherches Océanographiques de Dakar-Thiaroye (CRODT), Senegal and W. Ivantsoff (retired), Biology Science, Macquarie University NSW 2109, North Ryde, Australia iagnostic characters: Small, elongate fish, rarely exceeding 15 cm in length. Body elongate and Dsomewhat compressed. Short head, generally flattened dorsally, large eyes, sharp nose, mouth small, oblique and in terminal position, jaws subequal, reaching or slightly exceeding the anterior margin of the eye; premaxilla with ascending process of variable length, with lateral process present or absent; ramus of dentary bone elevated posteriorly or indistinct from anterior part of lower jaw; fine, small and sharp teeth on the jaws, on the roof of mouth (vomer, palatine, pterygoid) or on outside of mouth; 10 to 26 gill rakers long and slender on lower arm of first gill arch. Two well-separated dorsal fins, the first with 6 to 10 thin, flexible spines, located approximately in the middle of the body; the second dorsal and anal fins with a single small weak spine, 1 unbranched soft ray and a variable number of soft rays. Anal fin always originating slightly in advance of second dorsal fin; pectoral fins inserted high on the flanks, directly behind posterior rim of gill cover, with spine greatly reduced and first ray much thicker than those following. Abdomninal pelvic fins with 1 spine and 5 soft rays; forked caudal fin; anus away from the origin of the anal fin. Relatively large scales, cycloid (smooth). -
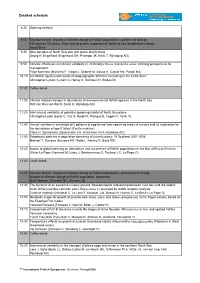
Schedule Onlinepdf
Detailed schedule 8:30 Opening session 9:00 Keynote lecture: Impacts of climate change on flatfish populations - patterns of change 100 days to 100 years: Short and long-term responses of flatfish to sea temperature change David Sims 9:30 Nine decades of North Sea sole and plaice distributions Georg H. Engelhard (Engelhard GH, Pinnegar JK, Kell LT, Rijnsdorp AD) 9:50 Climatic effects on recruitment variability in Platichthys flesus and Solea solea: defining perspectives for management. Filipe Martinho (Martinho F, Viegas I, Dolbeth M, Sousa H, Cabral HN, Pardal MA) 10:10 Are flatfish species with southern biogeographic affinities increasing in the Celtic Sea? Christopher Lynam (Lynam C, Harlay X, Gerritsen H, Stokes D) 10:30 Coffee break 11:00 Climate related changes in abundance of non-commercial flatfish species in the North Sea Ralf van Hal (van Hal R, Smits K, Rijnsdorp AD) 11:20 Inter-annual variability of potential spawning habitat of North Sea plaice Christophe Loots (Loots C, Vaz S, Koubii P, Planque B, Coppin F, Verin Y) 11:40 Annual variation in simulated drift patterns of egg/larvae from spawning areas to nursery and its implication for the abundance of age-0 turbot (Psetta maxima) Claus R. Sparrevohn (Sparrevohn CR, Hinrichsen H-H, Rijnsdorp AD) 12:00 Broadscale patterns in population dynamics of juvenile plaice: W Scotland 2001-2008 Michael T. Burrows (Burrows MT, Robb L, Harvey R, Batty RS) 12:20 Impact of global warming on abundance and occurrence of flatfish populations in the Bay of Biscay (France) Olivier Le Pape (Hermant -

Viral Haemorrhagic Septicaemia Virus (VHSV): on the Search for Determinants Important for Virulence in Rainbow Trout Oncorhynchus Mykiss
Downloaded from orbit.dtu.dk on: Nov 08, 2017 Viral haemorrhagic septicaemia virus (VHSV): on the search for determinants important for virulence in rainbow trout oncorhynchus mykiss Olesen, Niels Jørgen; Skall, H. F.; Kurita, J.; Mori, K.; Ito, T. Published in: 17th International Conference on Diseases of Fish And Shellfish Publication date: 2015 Document Version Publisher's PDF, also known as Version of record Link back to DTU Orbit Citation (APA): Olesen, N. J., Skall, H. F., Kurita, J., Mori, K., & Ito, T. (2015). Viral haemorrhagic septicaemia virus (VHSV): on the search for determinants important for virulence in rainbow trout oncorhynchus mykiss. In 17th International Conference on Diseases of Fish And Shellfish: Abstract book (pp. 147-147). [O-139] Las Palmas: European Association of Fish Pathologists. General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. DISCLAIMER: The organizer takes no responsibility for any of the content stated in the abstracts. -

Oryzias Latipes)
Betanodavirus infection in the freshwater model fish medaka (Oryzias latipes) Ryo Furusawa, Yasushi Okinaka,* and Toshihiro Nakai Graduate School of Biosphere Science, Hiroshima University, Higashi-hiroshima 739- 8528, Japan æfAuthor for correspondence: Yasushi Okinaka. Telephone: +8 1-82-424-7978. Fax: +81- 82-424-79 1 6. E-mail: [email protected] Running title: Betanodavirus infection in medaka Key words: medaka, betanodavirus, model fish, model virus, freshwater fish Total number of words; text (3532 words), summary (230 words) Total number of figures; 6 figures Total number of tables; 0 table SUMMARY Betanodaviruses, the causal agents of viral nervous necrosis in marine fish, have bipartite positive-sense RNA genomes. Because the genomes are the smallest and simplest among viruses, betanodaviruses are well studied using a genetic engineering system as model viruses, like the cases with the insect viruses, alphanodaviruses, the other members of the family Nodaviridae. However, studies of virus-host interactions have been limited because betanodaviruses basically infect marine fish at early developmental stages (larval and juvenile). These fish are only available for a few months of the year and are not suitable for the construction of a reversed genetics system. To overcome these problems, several freshwater fish species were tested for their susceptibility to betanodaviruses. We have demonstrated that adult medaka (Oryzias latipes), a well-known model fish, is susceptible to both Stripedjack nervous necrosis virus (the type species of the betanodaviruses) and Redspotted grouper nervous necrosis virus which have different host specificity in marine fish species. Infected medaka exhibited erratic swimming and the viruses were specifically localized to the brain, spinal cord, and retina of the infected fish, similar to the pattern of infection in naturally infected marine fish. -

Global Patterns of Ranavirus Detections
NOTE Global patterns of ranavirus detections Jesse L. Brunnera*, Deanna H. Olsonb, Matthew J. Grayc, Debra L. Millerd, and Amanda L.J. Duffuse aSchool of Biological Sciences, Washington State University, Pullman, WA 99164-4236, USA; bUSDA Forest Service, Pacific Northwest Research Station, Corvallis, OR 97331-8550, USA; cDepartment of Forestry, Wildlife and Fisheries, University of Tennessee Institute of Agriculture, Knoxville, TN 37996-4563, USA; dCollege of Veterinary Medicine, University of Tennessee Institute of Agriculture, Knoxville, TN 37996-4563, USA; eDepartment of Natural Sciences, Gordon State College, Barnesville, GA 30204, USA *[email protected] Abstract Ranaviruses are emerging pathogens of poikilothermic vertebrates. In 2015 the Global Ranavirus Reporting System (GRRS) was established as a centralized, open access, online database for reports of the presence (and absence) of ranavirus around the globe. The GRRS has multiple data layers (e.g., location, date, host(s) species, and methods of detection) of use to those studying the epidemiol- ogy, ecology, and evolution of this group of viruses. Here we summarize the temporal, spatial, diag- nostic, and host-taxonomic patterns of ranavirus reports in the GRRS. The number, distribution, and host diversity of ranavirus reports have increased dramatically since the mid 1990s, presumably in response to increased interest in ranaviruses and the conservation of their hosts, and also the availability of molecular diagnostics. Yet there are clear geographic and taxonomic biases among the OPEN ACCESS reports. We encourage ranavirus researchers to add their studies to the portal because such collation can provide collaborative opportunities and unique insights to our developing knowledge of this For personal use only. -

R M , July1979 Rum, Jlilio1979
FA0 Fisheriee Circular No. 706 FIR/C706 FA0 Cimulaire mur lee p8ohes No 706 FAO, CirouLerem de Pom~fbNo 706 SELECTED BIBLIOUUPHY ON PELAGIC FISH EGG AND LARVA SURVEYS BIBLIOWHIE SELECTIVE SUR LES PROSPECTIONS D'OEUFS ET DE LAFNFS DE POISSONS PELAGIQUES BIBLIOWA SELECCIONADA SOBRE RECONOCIMIEN'IQS DE HUEVOS Y LARVAS DE PECES PEZAOICOS Prepared by/Prdparge par/Preparada por Paul E. Smith Southwest Fisheries Center La Jolla, California, U.S.A. t /Y Sally L. Richardem Oregon State University Corvallis, Oregon, U.S.A. FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS ORGANISATION DES NATIONS UNIES POUR L'ALIMZNTATIW ET L'AaRICUL'NRE ORC;BFIZACI(YN DE LAS NACI- UNIDAS PAR4 LA AaRICUL"RA Y LA ALIMENTACION Rm, July1979 Ram, juillet 1979 Rum, jlilio1979 -1- 1. SCOPE, COVERAGE AND ORGAMI~TION This bibliography is intended to provide aocass to published information on ichthyo- plankton survey methods, identification of fish -,and larvae, and results of meyn thet have been carried out in the put. Although the bibliograpb in selective, its coverage in- cludes all published works through 1973, and it htu been extended by the addition of all papers resented at the Oban Symposium and publinhed in the eJwposiun proceedings (Blazter, J.H.S. red.) 1974. The early life history of firh. SpringarcVerlsg, Berlin). The fint five seotiau of this bibliomp4y list worh by name and drte anly aooordinn to subject category. Section 2 containe refer.noe8 on survey equipment and mothods. Section 3 includes descriptions of early life rtages organized by taxonomic group. Section 4 lists references on species identification of fish aggm and larva4 by region (specifically, by FA0 etatietical area). -

Virus Particle Structures
Virus Particle Structures Virus Particle Structures Palmenberg, A.C. and Sgro, J.-Y. COLOR PLATE LEGENDS These color plates depict the relative sizes and comparative virion structures of multiple types of viruses. The renderings are based on data from published atomic coordinates as determined by X-ray crystallography. The international online repository for 3D coordinates is the Protein Databank (www.rcsb.org/pdb/), maintained by the Research Collaboratory for Structural Bioinformatics (RCSB). The VIPER web site (mmtsb.scripps.edu/viper), maintains a parallel collection of PDB coordinates for icosahedral viruses and additionally offers a version of each data file permuted into the same relative 3D orientation (Reddy, V., Natarajan, P., Okerberg, B., Li, K., Damodaran, K., Morton, R., Brooks, C. and Johnson, J. (2001). J. Virol., 75, 11943-11947). VIPER also contains an excellent repository of instructional materials pertaining to icosahedral symmetry and viral structures. All images presented here, except for the filamentous viruses, used the standard VIPER orientation along the icosahedral 2-fold axis. With the exception of Plate 3 as described below, these images were generated from their atomic coordinates using a novel radial depth-cue colorization technique and the program Rasmol (Sayle, R.A., Milner-White, E.J. (1995). RASMOL: biomolecular graphics for all. Trends Biochem Sci., 20, 374-376). First, the Temperature Factor column for every atom in a PDB coordinate file was edited to record a measure of the radial distance from the virion center. The files were rendered using the Rasmol spacefill menu, with specular and shadow options according to the Van de Waals radius of each atom. -

Download Book (PDF)
e · ~ e t · aI ' A Field Guide to Grouper and Snapper Fishes of Andaman and Nicobar Islands (Family: SERRANIDAE, Subfamily: EPINEPHELINAE and Family: LUTJANIDAE) P. T. RAJAN Andaman & Nicobar Regional Station Zoological Survey of India Haddo, Port Blair - 744102 Edited by the Director, Zoological Survey of India, Kolkata Zoological Survey of India Kolkata CITATION Rajan, P. T. 2001. Afield guide to Grouper and Snapper Fishes of Andaman and Nicobar Islands. (Published - Director, Z.5.1.) Published : December, 2001 ISBN 81-85874-40-9 Front cover: Roving Coral Grouper (Plectropomus pessuliferus) Back cover : A School of Blue banded Snapper (Lutjanus lcasmira) © Government of India, 2001 ALL RIGHTS RESERVED • No part of this publication may be reproduced, stored in a retrieval system or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording or otherwise without the prior permission of the publisher. • This book is sold subject to the condition that it shall not, by way of trade, be lent, re-sold, hired out or otherwise disposed of without the publisher'S consent, in any form of binding or cover other than that in which it is published. • The correct price of this publication is the price printed on this page. Any revised price indicated by a rubber stamp or by a sticker or by any other means is incorrect and should be unacceptable. PRICE Indian Rs. 400.00 Foreign $ 25; £ 20 Published at the Publication Division by the Director, Zoological Survey of India, 234/4, AJe Bose Road, 2nd MSO Building, (13th Floor), Nizam Palace, Calcutta-700 020 after laser typesetting by Computech Graphics, Calcutta 700019 and printed at Power Printers, New Delhi - 110002.