BMC Evolutionary Biology, 9(1)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi
Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 (http://www.accessscience.com/) Article by: Boschung, Herbert Department of Biological Sciences, University of Alabama, Tuscaloosa, Alabama. Gardiner, Brian Linnean Society of London, Burlington House, Piccadilly, London, United Kingdom. Publication year: 2014 DOI: http://dx.doi.org/10.1036/1097-8542.680400 (http://dx.doi.org/10.1036/1097-8542.680400) Content Morphology Euteleostei Bibliography Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi The most recent group of actinopterygians (rayfin fishes), first appearing in the Upper Triassic (Fig. 1). About 26,840 species are contained within the Teleostei, accounting for more than half of all living vertebrates and over 96% of all living fishes. Teleosts comprise 517 families, of which 69 are extinct, leaving 448 extant families; of these, about 43% have no fossil record. See also: Actinopterygii (/content/actinopterygii/009100); Osteichthyes (/content/osteichthyes/478500) Fig. 1 Cladogram showing the relationships of the extant teleosts with the other extant actinopterygians. (J. S. Nelson, Fishes of the World, 4th ed., Wiley, New York, 2006) 1 of 9 10/7/2015 1:07 PM Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 Morphology Much of the evidence for teleost monophyly (evolving from a common ancestral form) and relationships comes from the caudal skeleton and concomitant acquisition of a homocercal tail (upper and lower lobes of the caudal fin are symmetrical). This type of tail primitively results from an ontogenetic fusion of centra (bodies of vertebrae) and the possession of paired bracing bones located bilaterally along the dorsal region of the caudal skeleton, derived ontogenetically from the neural arches (uroneurals) of the ural (tail) centra. -

Crestfish Lophotus Lacepede (Giorna, 1809) and Scalloped Ribbonfish Zu Cristatus (Bonelli, 1819) in the Northern Coast of Sicily, Italy
ISSN: 0001-5113 ACTA ADRIAT., ORIGINAL SCIENTIFIC PAPER AADRAY 58(1): 137 - 146, 2017 Occurrence of two rare species from order Lampriformes: Crestfish Lophotus lacepede (Giorna, 1809) and scalloped ribbonfish Zu cristatus (Bonelli, 1819) in the northern coast of Sicily, Italy Fabio FALSONE1, Michele Luca GERACI1, Danilo SCANNELLA1, Charles Odilichukwu R. OKPALA1, Giovan Battista GIUSTO1, Mar BOSCH-BELMAR2, Salvatore GANCITANO1 and Gioacchino BONO1 1Institute for the Coastal Marine Environment, IAMC‑CNR, 91026 Mazara del Vallo, Sicily, Italy 2Consorzio Nazionale Interuniversitario per le Scienze del Mare (CoNISMa), Rome, Italy Corresponding author, e‑mail: [email protected] The bony fish Lophotus lacepede (Giorna, 1809) and Zu cristatus (Bonelli, 1819) are the two species rarely recorded within the Mediterranean basin, usually reported as accidentally captured in depth (mesopelagic) fishing operations. In the current work, we present the first record of L. lacepede and Z. cristatus in fishing catches from southwestern Tyrrhenian Sea. Moreover, in order to improve existent biological/ecological knowledge, some bio-related aspects such as feeding aspect, sexual maturity and age estimate have been discussed. Key words: crestfish, scalloped ribbonfish, meristic features, vertebrae, growth ring INTRODUCTION species of Lophotidae family, the L. lacepede inhabits the epipelagic zone, although it could The target species of this study (Lophotus also be recorded in most oceans from the surface lacepede and Zu cristatus) belong to Lophotidae up to 1000 m depth (HEEMSTRA, 1986; PALMER, (Bonaparte, 1845) and Trachipteridae (Swain- 1986; OLNEY, 1999). First record of this spe- son, 1839) families respectively, including the cies in the Mediterranean Basin was from the Lampriformes order (consisted of 7 families). -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

Fish-T1K (Transcriptomes of 1,000 Fishes) Project: Large-Scale Transcriptome Data for Fish Evolution Studies Ying Sun1,2*†, Yu Huang2†, Xiaofeng Li2†, Carole C
Sun et al. GigaScience (2016) 5:18 DOI 10.1186/s13742-016-0124-7 COMMENTARY Open Access Fish-T1K (Transcriptomes of 1,000 Fishes) Project: large-scale transcriptome data for fish evolution studies Ying Sun1,2*†, Yu Huang2†, Xiaofeng Li2†, Carole C. Baldwin3, Zhuocheng Zhou4, Zhixiang Yan5, Keith A. Crandall6, Yong Zhang2, Xiaomeng Zhao2, Min Wang2,7, Alex Wong8, Chao Fang2, Xinhui Zhang2, Hai Huang9, Jose V. Lopez10, Kirk Kilfoyle10, Yong Zhang1, Guillermo Ortí6*, Byrappa Venkatesh11* and Qiong Shi2,7,12* Abstract Ray-finned fishes (Actinopterygii) represent more than 50 % of extant vertebrates and are of great evolutionary, ecologic and economic significance, but they are relatively underrepresented in ‘omics studies. Increased availability of transcriptome data for these species will allow researchers to better understand changes in gene expression, and to carry out functional analyses. An international project known as the “Transcriptomes of 1,000 Fishes” (Fish-T1K) project has been established to generate RNA-seq transcriptome sequences for 1,000 diverse species of ray-finned fishes. The first phase of this project has produced transcriptomes from more than 180 ray-finned fishes, representing 142 species and covering 51 orders and 109 families. Here we provide an overview of the goals of this project and the work done so far. Keywords: Fish-T1K, Fish, Transcriptome, RNA, Database, Biodiversity Background whole genomes of only 38 fish species have been pub- Ray-finned fishes (Actinopterygii) are the most diverse lished (Additional file 1) and, although the number is and abundant group of extant vertebrates. Thus far, ap- growing (Additional file 2), searching the National Cen- proximately 32,900 fish species are recorded in FishBase ter for Biotechnology Information (NCBI)’s Sequence [1]. -

Euteleostei: Aulopiformes) and the Timing of Deep-Sea Adaptations ⇑ Matthew P
Molecular Phylogenetics and Evolution 57 (2010) 1194–1208 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Estimating divergence times of lizardfishes and their allies (Euteleostei: Aulopiformes) and the timing of deep-sea adaptations ⇑ Matthew P. Davis a, , Christopher Fielitz b a Museum of Natural Science, Louisiana State University, 119 Foster Hall, Baton Rouge, LA 70803, USA b Department of Biology, Emory & Henry College, Emory, VA 24327, USA article info abstract Article history: The divergence times of lizardfishes (Euteleostei: Aulopiformes) are estimated utilizing a Bayesian Received 18 May 2010 approach in combination with knowledge of the fossil record of teleosts and a taxonomic review of fossil Revised 1 September 2010 aulopiform taxa. These results are integrated with a study of character evolution regarding deep-sea evo- Accepted 7 September 2010 lutionary adaptations in the clade, including simultaneous hermaphroditism and tubular eyes. Diver- Available online 18 September 2010 gence time estimations recover that the stem species of the lizardfishes arose during the Early Cretaceous/Late Jurassic in a marine environment with separate sexes, and laterally directed, round eyes. Keywords: Tubular eyes have arisen independently at different times in three deep-sea pelagic predatory aulopiform Phylogenetics lineages. Simultaneous hermaphroditism evolved a single time in the stem species of the suborder Character evolution Deep-sea Alepisauroidei, the clade of deep-sea aulopiforms during the Early Cretaceous. This result indicates the Euteleostei oldest known evolutionary event of simultaneous hermaphroditism in vertebrates, with the Alepisauroidei Hermaphroditism being the largest vertebrate clade with this reproductive strategy. Divergence times Ó 2010 Elsevier Inc. -
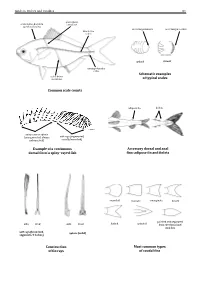
Field Identification Guide to the Living Marine Resources in Kenya
Guide to Orders and Families 81 lateral line scales above scales before dorsal fin outer margin smooth outer margin toothed (predorsal scales) lateral–line 114 scales cycloid ctenoidِّ scales circumpeduncular Schematic examples lateral line of typical scales scales below Common scale counts adipose fin finlets soft rays (segmented, spinyunbranched) rays or spines usually branched) (unsegmented, always Example of a continuous Accessory dorsal and anal dorsal fin of a spiny–rayed fish fins: adipose fin and finlets rounded truncate emarginate lunate side front side front from the dorsal and pointed and separated forked pointed soft rays (branched, spines (solid) segments, 2 halves) anal fins Construction Most common types of fin rays of caudal fins 82 Bony Fishes GUIDE TO ORDERS AND FAMILIES Order ELOPIFORMES – Tarpons and allies Fin spines absent; a single dorsal fin located above middle of body; pelvic fins in abdominal position; lateral line present; 23–25 branchiostegal rays; upper jaw extending past eye; tip of snout not overhanging mouth; colour silvery. ELOPIDAE Page 121 very small scales Ladyfishes To 90 cm. Coastal marine waters and estuaries; pelagic. A single species included in the Guide to Species.underside of head large mouth gular plate MEGALOPIDAE Page 121 last ray long Tarpons large scales To 55 cm. Coastal marine waters and estuaries; pelagic. A single species included in the Guide to Species.underside of head gular plate Order ALBULIFORMES – Bonefishes Fin spines absent; a single dorsal fin located above middle of body; pelvic fins in abdominal position; lateral line present; 6–16 branchiostegal rays; upper jaw not extending as far as front of eye; tip of snout overhanging mouth; colour silvery. -

DEEPFISHMAN Document 5 : Review of Parasites, Pathogens
DEEPFISHMAN Management And Monitoring Of Deep-sea Fisheries And Stocks Project number: 227390 Small or medium scale focused research action Topic: FP7-KBBE-2008-1-4-02 (Deepsea fisheries management) DEEPFISHMAN Document 5 Title: Review of parasites, pathogens and contaminants of deep sea fish with a focus on their role in population health and structure Due date: none Actual submission date: 10 June 2010 Start date of the project: April 1st, 2009 Duration : 36 months Organization Name of lead coordinator: Ifremer Dissemination Level: PU (Public) Date: 10 June 2010 Review of parasites, pathogens and contaminants of deep sea fish with a focus on their role in population health and structure. Matt Longshaw & Stephen Feist Cefas Weymouth Laboratory Barrack Road, The Nothe, Weymouth, Dorset DT4 8UB 1. Introduction This review provides a summary of the parasites, pathogens and contaminant related impacts on deep sea fish normally found at depths greater than about 200m There is a clear focus on worldwide commercial species but has an emphasis on records and reports from the north east Atlantic. In particular, the focus of species following discussion were as follows: deep-water squalid sharks (e.g. Centrophorus squamosus and Centroscymnus coelolepis), black scabbardfish (Aphanopus carbo) (except in ICES area IX – fielded by Portuguese), roundnose grenadier (Coryphaenoides rupestris), orange roughy (Hoplostethus atlanticus), blue ling (Molva dypterygia), torsk (Brosme brosme), greater silver smelt (Argentina silus), Greenland halibut (Reinhardtius hippoglossoides), deep-sea redfish (Sebastes mentella), alfonsino (Beryx spp.), red blackspot seabream (Pagellus bogaraveo). However, it should be noted that in some cases no disease or contaminant data exists for these species. -

Fishes-Of-The-Salish-Sea-Pp18.Pdf
NOAA Professional Paper NMFS 18 Fishes of the Salish Sea: a compilation and distributional analysis Theodore W. Pietsch James W. Orr September 2015 U.S. Department of Commerce NOAA Professional Penny Pritzker Secretary of Commerce Papers NMFS National Oceanic and Atmospheric Administration Kathryn D. Sullivan Scientifi c Editor Administrator Richard Langton National Marine Fisheries Service National Marine Northeast Fisheries Science Center Fisheries Service Maine Field Station Eileen Sobeck 17 Godfrey Drive, Suite 1 Assistant Administrator Orono, Maine 04473 for Fisheries Associate Editor Kathryn Dennis National Marine Fisheries Service Offi ce of Science and Technology Fisheries Research and Monitoring Division 1845 Wasp Blvd., Bldg. 178 Honolulu, Hawaii 96818 Managing Editor Shelley Arenas National Marine Fisheries Service Scientifi c Publications Offi ce 7600 Sand Point Way NE Seattle, Washington 98115 Editorial Committee Ann C. Matarese National Marine Fisheries Service James W. Orr National Marine Fisheries Service - The NOAA Professional Paper NMFS (ISSN 1931-4590) series is published by the Scientifi c Publications Offi ce, National Marine Fisheries Service, The NOAA Professional Paper NMFS series carries peer-reviewed, lengthy original NOAA, 7600 Sand Point Way NE, research reports, taxonomic keys, species synopses, fl ora and fauna studies, and data- Seattle, WA 98115. intensive reports on investigations in fi shery science, engineering, and economics. The Secretary of Commerce has Copies of the NOAA Professional Paper NMFS series are available free in limited determined that the publication of numbers to government agencies, both federal and state. They are also available in this series is necessary in the transac- exchange for other scientifi c and technical publications in the marine sciences. -

The Exceptional Diversity of Visual Adaptations in Deep-Sea Teleost Fishes
Seminars in Cell and Developmental Biology xxx (xxxx) xxx–xxx Contents lists available at ScienceDirect Seminars in Cell & Developmental Biology journal homepage: www.elsevier.com/locate/semcdb Review The exceptional diversity of visual adaptations in deep-sea teleost fishes Fanny de Busserolles*, Lily Fogg, Fabio Cortesi, Justin Marshall Queensland Brain Institute, The University of Queensland, St Lucia, Queensland 4072, Australia ARTICLE INFO ABSTRACT Keywords: The deep-sea is the largest and one of the dimmest habitats on earth. In this extreme environment, every photon Deep-sea teleost counts and may make the difference between life and death for its inhabitants. Two sources of light are present Dim-light vision in the deep-sea; downwelling light, that becomes dimmer and spectrally narrower with increasing depth until Ocular adaptation completely disappearing at around 1000 m, and bioluminescence, the light emitted by animals themselves. Retina Despite these relatively dark and inhospitable conditions, many teleost fish have made the deep-sea their home, Opsin relying heavily on vision to survive. Their visual systems have had to adapt, sometimes in astonishing and Bioluminescence bizarre ways. This review examines some aspects of the visual system of deep-sea teleosts and highlights the exceptional diversity in both optical and retinal specialisations. We also reveal how widespread several of these adaptations are across the deep-sea teleost phylogeny. Finally, the significance of some recent findings as well as the surprising diversity in visual adaptations is discussed. 1. Introduction or mate detection, to communicate, camouflage, or for navigation, in- cluding to stay within a particular depth range [4,5]. -
Marine Ecology Progress Series 503:23
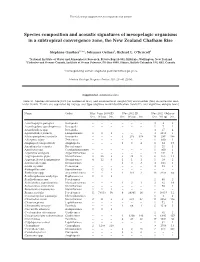
The following supplement accompanies the article 1 Species composition and acoustic signatures of mesopelagic organisms in a subtropical convergence zone, the New Zealand Chatham Rise Stéphane Gauthier1, 2,*, Johannes Oeffner1, Richard L. O’Driscoll1 1National Institute of Water and Atmospheric Research, Private Bag 14-901, Kilbirnie, Wellington, New Zealand 2Fisheries and Oceans Canada, Institute of Ocean Sciences, PO Box 6000, Sidney, British Columbia V8L 4B2, Canada *Corresponding author: [email protected] Marine Ecology Progress Series: 503: 23–40 (2014) Supplement. Additional data Table S1. Species occurrence (Occ.) in number of tows, and abundance in weight (Wt) and number (No.) in successful mid - water trawls. Trawls are separated by voyage and type (daytime mark identification trawls ID, and nighttime oblique tows) Name Order May−June 2008 ID Nov 2011 ID Nov 2011 Oblique Occ. Wt (g) No. Occ. Wt (g) No. Occ. Wt (g) No. Acanthephyra pelagica Decapoda − − − − − − 3 9 3 Acanthephyra quadrispinosa Decapoda − − − − − − 1 7 1 Acanthephyra spp. Decapoda − − − − − − 3 17 4 Agrostichthys parkeri Lampriformes 1 8 1 − − − 3 1813 3 Alainopasiphaea australis Decapoda − − − 3 273 356 14 287 366 Allocyttus niger Zeiformes − − − − − − 1 600 1 Amphipod (unspecified) Amphipoda − − − 1 3 4 8 34 17 Anoplogaster cornuta Beryciformes − − − − − − 1 25 1 Apristurus spp. Carcharhiniformes − − − − − − 1 900 1 Argentina elongata Argentiniformes − − − − − − 4 541 6 Argyropelecus gigas Stomiiformes 2 20 2 2 38 5 8 165 12 Argyropelecus hemigymnus Stomiiformes 4 12 6 1 1 1 1 19 2 Astronesthes spp. Stomiiformes − − − 1 3 3 3 103 3 Atolla wyvillei Coronatae − − − − − − 4 83 7 Bathophilus ater Stomiiformes 1 12 1 − − − − − − Bathylagus spp. -

Common Marine Species of Kanya Kumari District 1
COMMON MARINE SPECIES OF KANYA KUMARI DISTRICT 1. SARDINE Common name: Sardine Phylum: Chordata Class: Sardinella Order: Clupeiformes Scientific name: Sardinella longiceps Tamil Name: மத்鎿므ன் Sardines are silvery white coloured They are very small and elongated Spawning takes place during spring They feed on planktons They are rich in omega - 3 fatty acid, vitamin B – 12, proteins, trace elements (Ca, Mg, K, Zn, P) They are harvested during night time Uses Prevent heart diseases Prevents the risk of cancer Controls blood glucose level 2. TUNA Common name: Tuna Phylum: Chordata Class: Scombrinae Order: Scombriformes Scientific name: Thunnini Tamil Name:毂ரை므ன Elongated streamlined fish with round body They are metallic blue above and silvery below with iridescent shine Feeds on fishes, squid, shellfishes and planktons. Rich in oil, protein, fat, phosphorous, vitamin D omega - 3 fatty acid Spawning takes in spring Uses Lowers blood pressures Lowers cholesterol Relief from arthritis pain Reduce asthma 3. Edible Oyster Common name: Edible oyster Phylum:Mollusca Class: Bivalvia Order: Ostreida Scientific name: Crassostrea madrasensis Tamil Name:殿ப்ꮿ Sedentary bivalve with irregular shape (or) circular (or) elongated Filter feeder feeding planktons Rich in protein, glycogen and fat Spawning occurs in summer Uses Skin care Heart ailments Liver problems Blood pressure Water retention Pre menstrual tension 4. SEPIA Common name: Sepia Phylum:Mollusca Class: Cephalopoda Order: Sepiida Scientific name: Sepiida Tamil Name: கனவா므ன் They are large with ‘W’ shaped pupil They have 8 arms and 2 tentacles with suckers They eat small mollusc, crab, shrimp, worms They are rich in fat, cholesterol, sodium, protein and omega – 3 fatty acid Spawning takes place during summer & spring Uses Reduces cardiovascular diseases Obesity Hypertension High cholesterol Diabetes 5. -

Supplemental Fig S2.Pdf
hAT-38_HM_1p 1 Kolobok-1_Bf2p hAT-20_HM_1p 1 Kolobok-2_Bf2p hAT-47_SM_1p 0.96 Kolobok-1_CTe_2p 1 hAT-24_LCh_1p Kolobok-2_Xt2p hAT-29_LCh_1p 0.852 Kolobok-1_Aqu_2p Cyprinodontiformes Kolobok-4_DR_2p Perciformes Anoplopomatidae 1 Lampriformes Kolobok-1_AP_1p Pomacentridae 0.992 Kolobok-1_AP_2p 0.963 Holocentriformes 0.9 Kolobok-1_SKow_2p 0.985 Cypriniformes Kolobok-1_NV2p Kolobok-12_CGi_2p Labriformes 0.848 Kolobok-2_CGi_2p Perciformes Notothenioidei Gobiiformes 0.737 Siluriformes Perciformes Serranidae Cypriniformes Spariformes 1 Perciformes Percidae 0.992 Perciformes Sebastidae Esociformes 0.874 Holocentriformes Centrachiformes Anguilliformes Salmoniformes Gadiformes 1 Cyprinodontiformes Polypteryformes 0.937 0.999 Perciformes Serranidae Cypriniformes 0.999 0.997 0.9 Gadiformes Scombriformes Holocentriformes Perciformes Anoplopomatidae * Esociformes Beryciformes 0.805 Cyprinodontiformes Aulopiformes 0.9 Gadiformes 0.924 Perciformes Liparidae Clupeiformes Anoplopomtidae (1/1) Anoplopoma fimbria Gadiformes Cypriniformes Mugilidae 0.82 Cyprinodontiformes Percidae (1/6) hAT (863 AA) (863 hAT Clupeiformes 0.999 Etheostoma spectabi Anoplopomtidae (1/1) Perciformes Percidae Gadiformes Kolobok (137 AA) (137 Kolobok Anoplopoma fimbria Serranidae (3/8) Pseudochromidae 0.923 Epinephelus coioides Trachichthyiformes Gadiformes Percidae (4/6) Scombriformes E. lanceolatus Centrachiformes Sander vitreus Gadiformes E. moara Sander lucioperca Spariformes 0.995 Cypriniformes Sebastidae (9/9) Perca flavescens Sebastes aleutianus Perca fluviatilis Moronidae