Global Response of Saccharomyces Cerevisiae to an Alkylating Agent
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Critical Evaluation of Gene Expression Changes in Human Tissues In
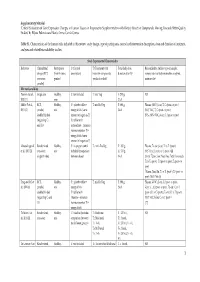
Supplementary Material ‘Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies’ by Biljana Pokimica and María-Teresa García-Conesa Table S1. Characteristics of the human trials included in this review: study design, type of participants, control and intervention description, dose and duration of treatment, analyses and related bioavailability studies. Study Experimental Characteristics Reference Clinical trial Participants C (Control T (Treatment with Total daily dose, Bioavailability studies: type of sample, design (RCT, (health status, description) bioactive compounds, duration (d or h)1 compounds and (or) metabolites analysed, crossover, gender) products or diet) main results2 parallel) Mix meals and diets Persson I et al., Single arm Healthy, C: not included T: mix Veg T: 250 g, NR 2000 [1] men 21 d Møller P et al., RCT, Healthy, C1: placebo tablet + T: mix FruVeg T: 600 g, Plasma: (NS↑) β-car, T, C2 (post- vs pre-) 2003 [2] parallel, mix energy drink (same 24 d (NC) VitC, T, C2 (post- vs pre-) double blinded amount of sugars as T) (NS↓, 69%) VitC, β-car, C1 (post- vs pre-) (regarding C1 C2: tablet with and C2) antioxidants + minerals (same amount as T) + energy drink (same amount of sugars as T) Almendingen K Randomized, Healthy, C: no proper control T1,2: mix FruVeg T1: 300 g, Plasma: ↑α-car, β-car, T2 vs T1 (post-) et al., 2005 [3] crossover, mix included (comparison T2: 750 g, (NS↑) Lyc, Lut, T2 vs T1 (post-) [4] single -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000238675-HmpCyc: Bacillus smithii 7_3_47FAA Cellular Overview Connections between pathways are omitted for legibility. -

Supporting Information
Supporting Information Figure S1. The functionality of the tagged Arp6 and Swr1 was confirmed by monitoring cell growth and sensitivity to hydeoxyurea (HU). Five-fold serial dilutions of each strain were plated on YPD with or without 50 mM HU and incubated at 30°C or 37°C for 3 days. Figure S2. Localization of Arp6 and Swr1 on chromosome 3. The binding of Arp6-FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) are compared. The position of Tel 3L, Tel 3R, CEN3, and the RP gene are shown under the panels. Figure S3. Localization of Arp6 and Swr1 on chromosome 4. The binding of Arp6-FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) in the whole chromosome region are compared. The position of Tel 4L, Tel 4R, CEN4, SWR1, and RP genes are shown under the panels. Figure S4. Localization of Arp6 and Swr1 on the region including the SWR1 gene of chromosome 4. The binding of Arp6- FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) are compared. The position and orientation of the SWR1 gene is shown. Figure S5. Localization of Arp6 and Swr1 on chromosome 5. The binding of Arp6-FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) are compared. The position of Tel 5L, Tel 5R, CEN5, and the RP genes are shown under the panels. Figure S6. Preferential localization of Arp6 and Swr1 in the 5′ end of genes. Vertical bars represent the binding ratio of proteins in each locus. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

The Distribution of Trehalase, Sucrase, Α-Amylase, Glucoamylase and Lactase
Downloaded from Br. J. Nutr. (197z),28, 129 https://www.cambridge.org/core The distribution of trehalase, sucrase, a-amylase, glucoamylase and lactase (8-galactosidase) along the small intestine of five pigs BY J. A. S'I'EVENS" AND D. E. KIDDER . IP address: Departments of Animal Husbandry and Veterinary Medicine, University of Bristol Langfwd House, Lungford, Bristol BSI 8 70U 170.106.40.40 (Received 24 September 1971 - Accepted 30 December 1971) I. Sucrase, trehalase (EC 3.2. I .28), a-amylase (RC 3.2.I. I) and glucoamylase ("-1.4 , on glucan glucohydrolase, BC 3.2.I .3) activities have been measured in the small intestine 25 Sep 2021 at 22:09:31 mucosa of five pigs varying in age from 19-30 weeks. The determinations were made at frequent intervals along the entire length starting 5 x 10-l m from the pylorus. Lactasc (/I-galactosidase, EC 3.2,I .23) has similarly been measured in one pig. 2. All these enzymes were present in the sample obtained from nearest to the pylorus and rose rapidly in the first few metres. 3. Trehalase and lactase were similarly distributed with a peak activity in the proximal quarter of the small intestine, falling to very low levels in the distal half. 4. Sucrase and glucoamylase resembled one another in distribution pattern with a peak , subject to the Cambridge Core terms of use, available at approximately midway along the small intestine, followed by a slight decrease in sucrase activity distally and a rather greater decrease with glucoamylase. 5. a-Amylasc activity, assumed to be duc to adsorhcd pancrcatic enzyme, had no regular pattern of distribution. -

Letters to Nature
letters to nature Received 7 July; accepted 21 September 1998. 26. Tronrud, D. E. Conjugate-direction minimization: an improved method for the re®nement of macromolecules. Acta Crystallogr. A 48, 912±916 (1992). 1. Dalbey, R. E., Lively, M. O., Bron, S. & van Dijl, J. M. The chemistry and enzymology of the type 1 27. Wolfe, P. B., Wickner, W. & Goodman, J. M. Sequence of the leader peptidase gene of Escherichia coli signal peptidases. Protein Sci. 6, 1129±1138 (1997). and the orientation of leader peptidase in the bacterial envelope. J. Biol. Chem. 258, 12073±12080 2. Kuo, D. W. et al. Escherichia coli leader peptidase: production of an active form lacking a requirement (1983). for detergent and development of peptide substrates. Arch. Biochem. Biophys. 303, 274±280 (1993). 28. Kraulis, P.G. Molscript: a program to produce both detailed and schematic plots of protein structures. 3. Tschantz, W. R. et al. Characterization of a soluble, catalytically active form of Escherichia coli leader J. Appl. Crystallogr. 24, 946±950 (1991). peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry 34, 3935±3941 29. Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and (1995). the thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281±296 (1991). 4. Allsop, A. E. et al.inAnti-Infectives, Recent Advances in Chemistry and Structure-Activity Relationships 30. Meritt, E. A. & Bacon, D. J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505± (eds Bently, P. H. & O'Hanlon, P. J.) 61±72 (R. Soc. Chem., Cambridge, 1997). -
Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_001431645Cyc: Stenotrophomonas panacihumi JCM 16536 Cellular Overview Connections between pathways are omitted for legibility. Anamika Kothari molybdate spermidine ammonium phosphate putrescine RS01245 RS09040 RS02020 RS09210 RS06445 ModC RS13295 RS07950 RS10935 RS05290 CcmE RS10150 RS15175 CcmA TatC RS15730 PotA FliR FliN FliF RS16780 RS13205 RS11350 RS08505 RS08615 RS04655 HtpX MreD RodA RS11935 SecA Amt RS14010 RS09060 RS11475 FliJ FtsE Cls FtsY SecF RS04370 RS14605 RS03760 RS08055 RS10260 RS10235 RS11315 RarD RS07635 RS12840 RS06270 RS15230 MurJ RS07085 RS02925 FlhA RS06365 AmpE MgtE RS05150 RS17505 PstB spermidine molybdate ammonium putrescine phosphate RS11800 FliD Cofactor, Carrier, and Vitamin Biosynthesis Amino Acid Degradation Aromatic Compound Macromolecule Modification tRNA-uridine 2-thiolation NADH repair (prokaryotes) Degradation di-trans,octa-cis UDP-N-acetyl- ditrans,octacis- a peptidoglycan with UDP-N-acetyl-α-D- L-alanyl-γ-D- an N-terminal- an N-terminal- a peptidoglycan (R)-4-hydroxy- -

Uracil and Beta-Alanine Degradation in Saccharomyces Kluyveri - Discovery of a Novel Catabolic Pathway
Downloaded from orbit.dtu.dk on: Sep 30, 2021 Uracil and beta-alanine degradation in Saccharomyces Kluyveri - discovery of a novel catabolic pathway Andersen, Gorm Publication date: 2006 Document Version Publisher's PDF, also known as Version of record Link back to DTU Orbit Citation (APA): Andersen, G. (2006). Uracil and beta-alanine degradation in Saccharomyces Kluyveri - discovery of a novel catabolic pathway. Technical University of Denmark. General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. Users may download and print one copy of any publication from the public portal for the purpose of private study or research. You may not further distribute the material or use it for any profit-making activity or commercial gain You may freely distribute the URL identifying the publication in the public portal If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Uracil and beta-alanine degradation in Saccharomyces kluyveri - Discovery of a novel catabolic pathway Ph.D. Thesis Gorm Andersen BioCentrum-DTU Technical University of Denmark May 2006 PREFACE The thesis presented here is a part of the requirements for the Ph.D. degree under the ”Chemistry and Biotechnology Programme” at the Technical University of Denmark (DTU). Since October 2002, I have been enrolled at BioCentrum-DTU. The study was financed by DTU and has been carried out under supervision of former Associate Professor at BioCentrum-DTU, (now Professor at Department of Cell and Organism Biology, Lund University, Sweden) Jure Piskur. -

Study on Yeast Enzymes Urc1p and Urc4p in a Novel Uracil Catabolic Pathway (URC)
Study on yeast enzymes Urc1p and Urc4p in a novel uracil catabolic pathway (URC) Dineshkumar Kandasamy Degree project in molecular biotechnology, 2012 Examensarbete i molekylär bioteknik 45 hp till masterexamen, 2012 Biology Education Centre, Uppsala University, and Department of Biology, Lund University Supervisors: Jure Piškur and Anna Rasmussen ABSTRACT Purine and pyrimidine bases are central precursors of DNA and RNA and their intracellular concentration is balanced by three pathways- de novo, salvage and catabolic pathways. Uracil catabolism pathway has been found in several bacteria and in some fungi (including yeast). Seven genes, URC1-7 have been found to be involved in this novel pathway. There are two “unknown genes” in the yeast Saccharomyces (Lachancea) kluyveri, namely URC1 and URC4, which play a central role in this pathway and their exact function remains a mystery. In this project, two S. kluyveri genes, URC1&URC4, were over-expressed in the bacterial system and successfully purified. Our preliminary functional assay showed that uridine monophosphate (UMP) is a likely substrate for Urc1p at pH 7, 25ºC. It was shown clearly that both uracil and uridine were not the substrate for Urc1p.We tried to phosphorylate chemically synthesized ribosylurea (RU) using Drosophila melanogaster deoxyribonucleoside kinase and compared the activity between phosphorylated and non- phosphorylated RU at different conditions. Phosphorylated ribosylurea seemed to be a likely substrate for Urc4p at pH 7, 37ºC. Keywords: Uridine monophosphate (UMP), ribosylurea (RU), uracil catabolism. Study on yeast enzymes Urc1p and Urc4p in a novel uracil catabolic pathway (URC) Popular science summary Dineshkumar Kandasamy Purines and pyrimidines are important constituent of nucleic acids and they are called building blocks of DNA and RNA. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Characterization of Visceral and Subcutaneous Adipose Tissue
J. Perinat. Med. 2016; 44(7): 813–835 Shali Mazaki-Tovi*, Adi L. Tarca, Edi Vaisbuch, Juan Pedro Kusanovic, Nandor Gabor Than, Tinnakorn Chaiworapongsa, Zhong Dong, Sonia S. Hassan and Roberto Romero* Characterization of visceral and subcutaneous adipose tissue transcriptome in pregnant women with and without spontaneous labor at term: implication of alternative splicing in the metabolic adaptations of adipose tissue to parturition DOI 10.1515/jpm-2015-0259 groups (unpaired analyses) and adipose tissue regions Received July 27, 2015. Accepted October 26, 2015. Previously (paired analyses). Selected genes were tested by quantita- published online March 19, 2016. tive reverse transcription-polymerase chain reaction. Abstract Results: Four hundred and eighty-two genes were differ- entially expressed between visceral and subcutaneous Objective: The aim of this study was to determine gene fat of pregnant women with spontaneous labor at term expression and splicing changes associated with par- (q-value < 0.1; fold change > 1.5). Biological processes turition and regions (visceral vs. subcutaneous) of the enriched in this comparison included tissue and vascu- adipose tissue of pregnant women. lature development as well as inflammatory and meta- Study design: The transcriptome of visceral and abdomi- bolic pathways. Differential splicing was found for 42 nal subcutaneous adipose tissue from pregnant women at genes [q-value < 0.1; differences in Finding Isoforms using term with (n = 15) and without (n = 25) spontaneous labor Robust Multichip Analysis scores > 2] between adipose was profiled with the Affymetrix GeneChip Human Exon tissue regions of women not in labor. Differential exon 1.0 ST array. Overall gene expression changes and the dif- usage associated with parturition was found for three ferential exon usage rate were compared between patient genes (LIMS1, HSPA5, and GSTK1) in subcutaneous tissues.