Mouse Pkib Conditional Knockout Project (CRISPR/Cas9)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
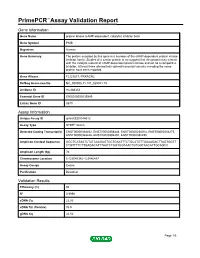
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name protein kinase (cAMP-dependent, catalytic) inhibitor beta Gene Symbol PKIB Organism Human Gene Summary The protein encoded by this gene is a member of the cAMP-dependent protein kinase inhibitor family. Studies of a similar protein in rat suggest that this protein may interact with the catalytic subunit of cAMP-dependent protein kinase and act as a competitive inhibitor. At least three alternatively spliced transcript variants encoding the same protein have been reported. Gene Aliases FLJ23817, PRKACN2 RefSeq Accession No. NC_000006.11, NT_025741.15 UniGene ID Hs.486354 Ensembl Gene ID ENSG00000135549 Entrez Gene ID 5570 Assay Information Unique Assay ID qHsaCED0044612 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000368452, ENST00000368448, ENST00000258014, ENST00000354275, ENST00000368446, ENST00000392491, ENST00000392490 Amplicon Context Sequence GGCTCATAATCTATCAAGAGTGCTGAATTTCTGCATGTTGAAAGACTTAGTGGTT CTGTTTTCTTGAGACATTTAATCTGGTGGTAACTGTGGTAACATTGCAGCC Amplicon Length (bp) 76 Chromosome Location 6:123046342-123046447 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 95 R2 0.9996 cDNA Cq 22.83 cDNA Tm (Celsius) 76.5 gDNA Cq 24.52 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report PKIB, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve -

Location Analysis of Estrogen Receptor Target Promoters Reveals That
Location analysis of estrogen receptor ␣ target promoters reveals that FOXA1 defines a domain of the estrogen response Jose´ e Laganie` re*†, Genevie` ve Deblois*, Ce´ line Lefebvre*, Alain R. Bataille‡, Franc¸ois Robert‡, and Vincent Gigue` re*†§ *Molecular Oncology Group, Departments of Medicine and Oncology, McGill University Health Centre, Montreal, QC, Canada H3A 1A1; †Department of Biochemistry, McGill University, Montreal, QC, Canada H3G 1Y6; and ‡Laboratory of Chromatin and Genomic Expression, Institut de Recherches Cliniques de Montre´al, Montreal, QC, Canada H2W 1R7 Communicated by Ronald M. Evans, The Salk Institute for Biological Studies, La Jolla, CA, July 1, 2005 (received for review June 3, 2005) Nuclear receptors can activate diverse biological pathways within general absence of large scale functional data linking these putative a target cell in response to their cognate ligands, but how this binding sites with gene expression in specific cell types. compartmentalization is achieved at the level of gene regulation is Recently, chromatin immunoprecipitation (ChIP) has been used poorly understood. We used a genome-wide analysis of promoter in combination with promoter or genomic DNA microarrays to occupancy by the estrogen receptor ␣ (ER␣) in MCF-7 cells to identify loci recognized by transcription factors in a genome-wide investigate the molecular mechanisms underlying the action of manner in mammalian cells (20–24). This technology, termed 17-estradiol (E2) in controlling the growth of breast cancer cells. ChIP-on-chip or location analysis, can therefore be used to deter- We identified 153 promoters bound by ER␣ in the presence of E2. mine the global gene expression program that characterize the Motif-finding algorithms demonstrated that the estrogen re- action of a nuclear receptor in response to its natural ligand. -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

Molecular Effects of Isoflavone Supplementation Human Intervention Studies and Quantitative Models for Risk Assessment
Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis committee Promotors Prof. Dr Pieter van ‘t Veer Professor of Nutritional Epidemiology Wageningen University Prof. Dr Evert G. Schouten Emeritus Professor of Epidemiology and Prevention Wageningen University Co-promotors Dr Anouk Geelen Assistant professor, Division of Human Nutrition Wageningen University Dr Lydia A. Afman Assistant professor, Division of Human Nutrition Wageningen University Other members Prof. Dr Jaap Keijer, Wageningen University Dr Hubert P.J.M. Noteborn, Netherlands Food en Consumer Product Safety Authority Prof. Dr Yvonne T. van der Schouw, UMC Utrecht Dr Wendy L. Hall, King’s College London This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 20 June 2014 at 13.30 p.m. in the Aula. Vera van der Velpen Molecular effects of isoflavone supplementation: Human intervention studies and quantitative models for risk assessment 154 pages PhD thesis, Wageningen University, Wageningen, NL (2014) With references, with summaries in Dutch and English ISBN: 978-94-6173-952-0 ABSTRact Background: Risk assessment can potentially be improved by closely linked experiments in the disciplines of epidemiology and toxicology. -

75 2. INTRODUCTION Triple-Negative Breast Cancer (TNBC)
[Frontiers in Bioscience, Scholar, 11, 75-88, March 1, 2019] The persisting puzzle of racial disparity in triple negative breast cancer: looking through a new lens Chakravarthy Garlapati1, Shriya Joshi1, Bikram Sahoo1, Shobhna Kapoor2, Ritu Aneja1 1Department of Biology, Georgia State University, Atlanta, GA, USA, 2Department of Chemistry, Indian Institute of Technology Bombay, Powai, India TABLE OF CONTENTS 1. Abstract 2. Introduction 3. Dissecting the TNBC racially disparate burden 3.1. Does race influence TNBC onset and progression? 3.2. Tumor microenvironment in TNBC and racial disparity 3.3. Differential gene signatures and pathways in racially distinct TNBC 3.4. Our Perspective: Looking racial disparity through a new lens 4. Conclusion 5. Acknowledgement 6. References 1. ABSTRACT 2. INTRODUCTION Triple-negative breast cancer (TNBC) Triple-negative breast cancer (TNBC), is characterized by the absence of estrogen a subtype of breast cancer (BC), accounts for and progesterone receptors and absence 15-20% of all BC diagnoses in the US. It has of amplification of human epidermal growth been recognized that women of African descent factor receptor (HER2). This disease has no are twice as likely to develop TNBC than approved treatment with a poor prognosis women of European descent (1). As the name particularly in African-American (AA) as foretells, TNBCs lack estrogen, progesterone, compared to European-American (EA) and human epidermal growth factor receptors. patients. Gene ontology analysis showed Unfortunately, TNBCs are defined by what they specific gene pathways that are differentially “lack” rather than what they “have” and thus this regulated and gene signatures that are negative nomenclature provides no actionable differentially expressed in AA as compared to information on “druggable” targets. -

Integrated Analysis of Gene Expression Changes Associated
Miao et al. Lipids in Health and Disease (2019) 18:92 https://doi.org/10.1186/s12944-019-1032-5 RESEARCH Open Access Integrated analysis of gene expression changes associated with coronary artery disease Liu Miao1 , Rui-Xing Yin1,2,3* , Feng Huang1,2,3, Shuo Yang1, Wu-Xian Chen1 and Jin-Zhen Wu1 Abstract Background: This study investigated the pathways and genes involved in coronary artery disease (CAD) and the associated mechanisms. Methods: Two array data sets of GSE19339 and GSE56885 were downloaded. The limma package was used to analyze the differentially expressed genes (DEGs) in normal and CAD specimens. Examination of DEGs through Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and Gene Ontology annotation was achieved by Database for Annotation, Visualization and Integrated Discovery (DAVID). The Cytoscape software facilitated the establishment of the protein-protein interaction (PPI) network and Molecular Complex Detection (MCODE) was performed for the significant modules. Results: We identified 413 DEGs (291 up-regulated and 122 down-regulated). Approximately 256 biological processes, only 1 cellular component, and 21 molecular functions were identified by GO analysis and 10 pathways were enriched by KEGG. Moreover, 264 protein pairs and 64 nodes were visualized by the PPI network. After the MCODE analysis, the top 4 high degree genes, including interleukin 1 beta (IL1B, degree = 29), intercellular adhesion molecule 1 (ICAM1, degree = 25), Jun proto-oncogene (JUN, degree = 23) and C-C motif chemokine ligand 2 (CCL2, degree = 20) had been identified to validate in RT-PCR and Cox proportional hazards regression between CAD and normals. Conclusions: The relative expression of IL1B, ICAM1 and CCL2 was higher in CAD than in normal controls (P <0.05–0. -

Global Identification of MLL2-Targeted Loci Reveals Mll2ts Role in Diverse
Global identification of MLL2-targeted loci reveals MLL2’s role in diverse signaling pathways Changcun Guoa,b,c,1, Chun-Chi Changa,b,c,1, Matthew Worthama,b,c, Lee H. Chena,b,c, Dawn N. Kernagisc, Xiaoxia Qind, Young-Wook Choe,2, Jen-Tsan Chid, Gerald A. Granta,b,f, Roger E. McLendona,b,c, Hai Yana,b,c, Kai Gee, Nickolas Papadopoulosg, Darell D. Bignera,b,c, and Yiping Hea,b,c,3 aThe Preston Robert Tisch Brain Tumor Center at Duke, bPediatric Brain Tumor Foundation Institute, cDepartment of Pathology, dInstitute for Genome Sciences and Policy, and fDepartment of Surgery, Duke University, Durham, NC 27710; eLaboratory of Endocrinology and Receptor Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892; and gLudwig Center for Cancer Genetics and Therapeutics, Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD 21231 Edited* by Bert Vogelstein, Johns Hopkins University, Baltimore, MD, and approved September 14, 2012 (received for review June 1, 2012) Myeloid/lymphoid or mixed-lineage leukemia (MLL)-family genes SWI/SNF, the MLL2 or MLL3 (hereafter referred to as MLL2/ encode histone lysine methyltransferases that play important 3) complex has been found to play essential roles as a coactivator roles in epigenetic regulation of gene transcription. MLL genes for transcriptional activation by nuclear hormone receptors (11). are frequently mutated in human cancers. Unlike MLL1, MLL2 (also Consistent with this notion, previous studies have shown that known as ALR/MLL4) and its homolog MLL3 are not well-under- MLL2/3 complexes regulate Hox gene transcription in an es- stood. -

Diverse Gene Expression and DNA Methylation Profiles Correlate with Differential Adaptation of Breast Cancer Cells to the Antiestrogens Tamoxifen and Fulvestrant
Research Article Diverse Gene Expression and DNA Methylation Profiles Correlate with Differential Adaptation of Breast Cancer Cells to the Antiestrogens Tamoxifen and Fulvestrant Meiyun Fan,1 Pearlly S. Yan,2 Cori Hartman-Frey,1 Lei Chen,1 Henry Paik,1 Samuel L. Oyer,1 Jonathan D. Salisbury,1 Alfred S.L. Cheng,2 Lang Li,3 Phillip H. Abbosh,1 Tim H-M. Huang,2 and Kenneth P. Nephew1,4,5 1Medical Sciences, Indiana University School of Medicine, Bloomington, Indiana; 2Division of Human Cancer Genetics, Comprehensive Cancer Center, Ohio State University, Columbus, Ohio; 3Division of Biostatistics, Department of Medicine and 4Department of Cellular and Integrative Physiology, Indiana University School of Medicine; and 5Indiana University Cancer Center, Indianapolis, Indiana Abstract Introduction The development of targeted therapies for antiestrogen- The steroid hormone estrogen is strongly implicated in the resistant breast cancer requires a detailed understanding development and progression of breast cancer (1). The primary of its molecular characteristics. To further elucidate the mediator of estrogen action in breast cancer cells is estrogen molecular events underlying acquired resistance to the receptor a (ERa), a ligand-activated transcription factor (1). antiestrogens tamoxifen and fulvestrant, we established Consequently, the leading drugs used for endocrine therapy of drug-resistant sublines from a single colony of hormone- breast cancer all block ERa activity, including antiestrogens (i.e., dependent breast cancer MCF7 cells. These model systems tamoxifen and fulvestrant) and aromatase inhibitors (2). Despite allowed us to examine the cellular and molecular changes the efficacy and favorable safety profile of these agents, the use of induced by antiestrogens in the context of a uniform clonal endocrine therapy is limited by the onset of drug resistance, in background. -

The Role of Natural Selection in Human Evolution – Insights from Latin America
Genetics and Molecular Biology, 39, 3, 302-311 (2016) Copyright © 2016, Sociedade Brasileira de Genética. Printed in Brazil DOI: http://dx.doi.org/10.1590/1678-4685-GMB-2016-0020 Review Article The role of natural selection in human evolution – insights from Latin America Francisco M. Salzano1 1Departamento de Genética, Instituto de Biociências, Universidade Federal do Rio Grande do Sul (UFRGS), Porto Alegre, RS, Brazil. Abstract A brief introduction considering Darwin’s work, the evolutionary synthesis, and the scientific biological field around the 1970s and subsequently, with the molecular revolution, was followed by selected examples of recent investiga- tions dealing with the selection-drift controversy. The studies surveyed included the comparison between essential genes in humans and mice, selection in Africa and Europe, and the possible reasons why females in humans remain healthy and productive after menopause, in contrast with what happens in the great apes. At the end, selected exam- ples of investigations performed in Latin America, related to the action of selection for muscle performance, acetylation of xenobiotics, high altitude and tropical forest adaptations were considered. Despite dissenting views, the influence of positive selection in a considerable portion of the human genome cannot presently be dismissed. Keywords: natural selection, human evolution, population genetics, human adaptation, history of genetics. Received: February 10, 2016; Accepted: May 8, 2016. History deniably indicated that we had derived from -

Functional Genomics Approach Identifies Novel Signaling Regulators of Tgfa Ectodomain Shedding
Published OnlineFirst October 10, 2017; DOI: 10.1158/1541-7786.MCR-17-0140 Signal Transduction Molecular Cancer Research Functional Genomics Approach Identifies Novel Signaling Regulators of TGFa Ectodomain Shedding Jennifer L. Wilson1, Eirini Kefaloyianni2, Lauren Stopfer1, Christina Harrison3, Venkata S. Sabbisetti4, Ernest Fraenkel1,5, Douglas A. Lauffenburger1, and Andreas Herrlich2 Abstract Ectodomain shedding of cell-surface precursor proteins by shRNA genomic screen, computational network analysis, and metalloproteases generates important cellular signaling molecules. dedicated validation tests succeeded in identifying several key Of importance for disease is the release of ligands that activate the signaling pathways as novel regulators of TGFa shedding in cancer EGFR, such as TGFa, which is mostly carried out by ADAM17 [a cells. Most notably, a cluster of genes with NFkBpathwayregu- member of the A-disintegrin and metalloprotease (ADAM) latory functions was found to strongly influence TGFa release, domain family]. EGFR ligand shedding has been linked to many albeit independent of their NFkB regulatory functions. Inflamma- diseases, in particular cancer development, growth and metastasis, tory regulators thus also govern cancer cell growth–promoting as well as resistance to cancer therapeutics. Excessive EGFR ligand ectodomain cleavage, lending mechanistic understanding to the release can outcompete therapeutic EGFR inhibition or the inhi- well-known connection between inflammation and cancer. bition of other growth factor pathways by providing bypass signaling via EGFR activation. Drugging metalloproteases directly Implications: Using genomic screens and network analysis, this have failed clinically because it indiscriminately affected shedding study defines targets that regulate ectodomain shedding and of numerous substrates. It is therefore essential to identify regu- suggests new treatment opportunities for EGFR-driven cancers. -

Global Identification of MLL2-Targeted Loci Reveals Mll2ts Role in Diverse Signaling Pathways
Global identification of MLL2-targeted loci reveals MLL2’s role in diverse signaling pathways Changcun Guoa,b,c,1, Chun-Chi Changa,b,c,1, Matthew Worthama,b,c, Lee H. Chena,b,c, Dawn N. Kernagisc, Xiaoxia Qind, Young-Wook Choe,2, Jen-Tsan Chid, Gerald A. Granta,b,f, Roger E. McLendona,b,c, Hai Yana,b,c, Kai Gee, Nickolas Papadopoulosg, Darell D. Bignera,b,c, and Yiping Hea,b,c,3 aThe Preston Robert Tisch Brain Tumor Center at Duke, bPediatric Brain Tumor Foundation Institute, cDepartment of Pathology, dInstitute for Genome Sciences and Policy, and fDepartment of Surgery, Duke University, Durham, NC 27710; eLaboratory of Endocrinology and Receptor Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892; and gLudwig Center for Cancer Genetics and Therapeutics, Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD 21231 Edited* by Bert Vogelstein, Johns Hopkins University, Baltimore, MD, and approved September 14, 2012 (received for review June 1, 2012) Myeloid/lymphoid or mixed-lineage leukemia (MLL)-family genes SWI/SNF, the MLL2 or MLL3 (hereafter referred to as MLL2/ encode histone lysine methyltransferases that play important 3) complex has been found to play essential roles as a coactivator roles in epigenetic regulation of gene transcription. MLL genes for transcriptional activation by nuclear hormone receptors (11). are frequently mutated in human cancers. Unlike MLL1, MLL2 (also Consistent with this notion, previous studies have shown that known as ALR/MLL4) and its homolog MLL3 are not well-under- MLL2/3 complexes regulate Hox gene transcription in an es- stood. -

Characterization of Foxp2 Functions in the Mouse Cortex Vera Medvedeva
Characterization of Foxp2 functions in the mouse cortex Vera Medvedeva To cite this version: Vera Medvedeva. Characterization of Foxp2 functions in the mouse cortex. Neurons and Cognition [q-bio.NC]. Université Pierre et Marie Curie - Paris VI, 2015. English. NNT : 2015PA066118. tel- 01192592 HAL Id: tel-01192592 https://tel.archives-ouvertes.fr/tel-01192592 Submitted on 3 Sep 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Université Pierre et Marie Curie Ecole doctorale Cerveau Cognition Comportement Institut du Fer à Moulin / Neurodevelopmental disorders Characterization of Foxp2 functions in the mouse cortex Par Vera Pavlovna MEDVEDEVA Thèse de doctorat de Neurogenetics Dirigée par Matthias Groszer, CR1, Inserm Présentée et soutenue publiquement le 17 Juin 2015 Devant un jury composé de : Pr. Ann LOHOF Présidente Dr. Markus WOEHR Rapporteur Dr. Pierre BILLUART Rapporteur Pr. Josef PRILLER Examinateur Dr. Carlos PARRAS Examinateur Dédicace To my grandfather Leonid Michailovich Kolmak Леониду Михайловичу Колмаку Acknowledgements First of all I would like to thank my supervisor Matthias Groszer who has kindly given me the opportunity to work in his laboratory and pursue his and sometimes my own scientific ideas. The scientific and, even more, life experiences in his laboratory were truly exceptional and rather determining for me, due to the very special environment established there.