Functional Genomics Approach Identifies Novel Signaling Regulators of Tgfa Ectodomain Shedding
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Exceptional Conservation of Horse–Human Gene Order on X Chromosome Revealed by High-Resolution Radiation Hybrid Mapping
Exceptional conservation of horse–human gene order on X chromosome revealed by high-resolution radiation hybrid mapping Terje Raudsepp*†, Eun-Joon Lee*†, Srinivas R. Kata‡, Candice Brinkmeyer*, James R. Mickelson§, Loren C. Skow*, James E. Womack‡, and Bhanu P. Chowdhary*¶ʈ *Department of Veterinary Anatomy and Public Health, ‡Department of Veterinary Pathobiology, College of Veterinary Medicine, and ¶Department of Animal Science, College of Agriculture and Life Science, Texas A&M University, College Station, TX 77843; and §Department of Veterinary Pathobiology, University of Minnesota, 295f AS͞VM, St. Paul, MN 55108 Contributed by James E. Womack, December 30, 2003 Development of a dense map of the horse genome is key to efforts ciated with the traits, once they are mapped by genetic linkage aimed at identifying genes controlling health, reproduction, and analyses with highly polymorphic markers. performance. We herein report a high-resolution gene map of the The X chromosome is the most conserved mammalian chro- horse (Equus caballus) X chromosome (ECAX) generated by devel- mosome (18, 19). Extensive comparisons of structure, organi- oping and typing 116 gene-specific and 12 short tandem repeat zation, and gene content of this chromosome in evolutionarily -markers on the 5,000-rad horse ؋ hamster whole-genome radia- diverse mammals have revealed a remarkable degree of conser tion hybrid panel and mapping 29 gene loci by fluorescence in situ vation (20–22). Until now, the chromosome has been best hybridization. The human X chromosome sequence was used as a studied in humans and mice, where the focus of research has template to select genes at 1-Mb intervals to develop equine been the intriguing patterns of X inactivation and the involve- orthologs. -

1 Long-Read Genome Sequencing for the Diagnosis Of
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.02.185447; this version posted September 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Long-read genome sequencing for the diagnosis of neurodevelopmental disorders Susan M. Hiatt1, James M.J. Lawlor1, Lori H. Handley1, Ryne C. Ramaker1, Brianne B. Rogers1,2, E. Christopher Partridge1, Lori Beth Boston1, Melissa Williams1, Christopher B. Plott1, Jerry Jenkins1, David E. Gray1, James M. Holt1, Kevin M. Bowling1, E. Martina Bebin3, Jane Grimwood1, Jeremy Schmutz1, Gregory M. Cooper1* 1HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA, 35806 2Department of Genetics, University of Alabama at Birmingham, Birmingham, AL, USA, 35924 3Department of Neurology, University of Alabama at Birmingham, Birmingham, AL, USA, 35924 *[email protected], 256-327-9490 Conflicts of Interest The authors all declare no conflicts of interest. 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.07.02.185447; this version posted September 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Purpose Exome and genome sequencing have proven to be effective tools for the diagnosis of neurodevelopmental disorders (NDDs), but large fractions of NDDs cannot be attributed to currently detectable genetic variation. This is likely, at least in part, a result of the fact that many genetic variants are difficult or impossible to detect through typical short-read sequencing approaches. -
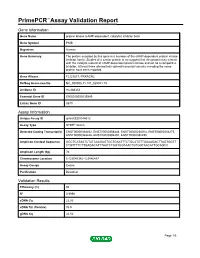
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name protein kinase (cAMP-dependent, catalytic) inhibitor beta Gene Symbol PKIB Organism Human Gene Summary The protein encoded by this gene is a member of the cAMP-dependent protein kinase inhibitor family. Studies of a similar protein in rat suggest that this protein may interact with the catalytic subunit of cAMP-dependent protein kinase and act as a competitive inhibitor. At least three alternatively spliced transcript variants encoding the same protein have been reported. Gene Aliases FLJ23817, PRKACN2 RefSeq Accession No. NC_000006.11, NT_025741.15 UniGene ID Hs.486354 Ensembl Gene ID ENSG00000135549 Entrez Gene ID 5570 Assay Information Unique Assay ID qHsaCED0044612 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000368452, ENST00000368448, ENST00000258014, ENST00000354275, ENST00000368446, ENST00000392491, ENST00000392490 Amplicon Context Sequence GGCTCATAATCTATCAAGAGTGCTGAATTTCTGCATGTTGAAAGACTTAGTGGTT CTGTTTTCTTGAGACATTTAATCTGGTGGTAACTGTGGTAACATTGCAGCC Amplicon Length (bp) 76 Chromosome Location 6:123046342-123046447 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 95 R2 0.9996 cDNA Cq 22.83 cDNA Tm (Celsius) 76.5 gDNA Cq 24.52 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report PKIB, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve -

Mouse Ppef1 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Ppef1 Knockout Project (CRISPR/Cas9) Objective: To create a Ppef1 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Ppef1 gene (NCBI Reference Sequence: NM_011147 ; Ensembl: ENSMUSG00000062168 ) is located on Mouse chromosome X. 16 exons are identified, with the ATG start codon in exon 1 and the TAA stop codon in exon 16 (Transcript: ENSMUST00000071204). Exon 2~4 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Homozygous null female and hemizygous null male mice are viable, fertile and display no overt abnormalities. Exon 2 starts from about 2.41% of the coding region. Exon 2~4 covers 18.41% of the coding region. The size of effective KO region: ~10105 bp. The KO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 3 4 16 Legends Exon of mouse Ppef1 Knockout region Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 2 is aligned with itself to determine if there are tandem repeats. No significant tandem repeat is found in the dot plot matrix. So this region is suitable for PCR screening or sequencing analysis. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section downstream of Exon 4 is aligned with itself to determine if there are tandem repeats. -

Location Analysis of Estrogen Receptor Target Promoters Reveals That
Location analysis of estrogen receptor ␣ target promoters reveals that FOXA1 defines a domain of the estrogen response Jose´ e Laganie` re*†, Genevie` ve Deblois*, Ce´ line Lefebvre*, Alain R. Bataille‡, Franc¸ois Robert‡, and Vincent Gigue` re*†§ *Molecular Oncology Group, Departments of Medicine and Oncology, McGill University Health Centre, Montreal, QC, Canada H3A 1A1; †Department of Biochemistry, McGill University, Montreal, QC, Canada H3G 1Y6; and ‡Laboratory of Chromatin and Genomic Expression, Institut de Recherches Cliniques de Montre´al, Montreal, QC, Canada H2W 1R7 Communicated by Ronald M. Evans, The Salk Institute for Biological Studies, La Jolla, CA, July 1, 2005 (received for review June 3, 2005) Nuclear receptors can activate diverse biological pathways within general absence of large scale functional data linking these putative a target cell in response to their cognate ligands, but how this binding sites with gene expression in specific cell types. compartmentalization is achieved at the level of gene regulation is Recently, chromatin immunoprecipitation (ChIP) has been used poorly understood. We used a genome-wide analysis of promoter in combination with promoter or genomic DNA microarrays to occupancy by the estrogen receptor ␣ (ER␣) in MCF-7 cells to identify loci recognized by transcription factors in a genome-wide investigate the molecular mechanisms underlying the action of manner in mammalian cells (20–24). This technology, termed 17-estradiol (E2) in controlling the growth of breast cancer cells. ChIP-on-chip or location analysis, can therefore be used to deter- We identified 153 promoters bound by ER␣ in the presence of E2. mine the global gene expression program that characterize the Motif-finding algorithms demonstrated that the estrogen re- action of a nuclear receptor in response to its natural ligand. -

Identification of Conserved Genes Triggering Puberty in European Sea
Blázquez et al. BMC Genomics (2017) 18:441 DOI 10.1186/s12864-017-3823-2 RESEARCHARTICLE Open Access Identification of conserved genes triggering puberty in European sea bass males (Dicentrarchus labrax) by microarray expression profiling Mercedes Blázquez1,2* , Paula Medina1,2,3, Berta Crespo1,4, Ana Gómez1 and Silvia Zanuy1* Abstract Background: Spermatogenesisisacomplexprocesscharacterized by the activation and/or repression of a number of genes in a spatio-temporal manner. Pubertal development in males starts with the onset of the first spermatogenesis and implies the division of primary spermatogonia and their subsequent entry into meiosis. This study is aimed at the characterization of genes involved in the onset of puberty in European sea bass, and constitutes the first transcriptomic approach focused on meiosis in this species. Results: European sea bass testes collected at the onset of puberty (first successful reproduction) were grouped in stage I (resting stage), and stage II (proliferative stage). Transition from stage I to stage II was marked by an increase of 11ketotestosterone (11KT), the main fish androgen, whereas the transcriptomic study resulted in 315 genes differentially expressed between the two stages. The onset of puberty induced 1) an up-regulation of genes involved in cell proliferation, cell cycle and meiosis progression, 2) changes in genes related with reproduction and growth, and 3) a down-regulation of genes included in the retinoic acid (RA) signalling pathway. The analysis of GO-terms and biological pathways showed that cell cycle, cell division, cellular metabolic processes, and reproduction were affected, consistent with the early events that occur during the onset of puberty. -

The Role of Cyclin B3 in Mammalian Meiosis
THE ROLE OF CYCLIN B3 IN MAMMALIAN MEIOSIS by Mehmet Erman Karasu A Dissertation Presented to the Faculty of the Louis V. Gerstner Jr. Graduate School of Biomedical Sciences, Memorial Sloan Kettering Cancer Center In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy New York, NY November, 2018 Scott Keeney, PhD Date Dissertation Mentor Copyright © Mehmet Erman Karasu 2018 DEDICATION I would like to dedicate this thesis to my parents, Mukaddes and Mustafa Karasu. I have been so lucky to have their support and unconditional love in this life. ii ABSTRACT Cyclins and cyclin dependent kinases (CDKs) lie at the center of the regulation of the cell cycle. Cyclins as regulatory partners of CDKs control the switch-like cell cycle transitions that orchestrate orderly duplication and segregation of genomes. Similar to somatic cell division, temporal regulation of cyclin-CDK activity is also important in meiosis, which is the specialized cell division that generates gametes for sexual production by halving the genome. Meiosis does so by carrying out one round of DNA replication followed by two successive divisions without another intervening phase of DNA replication. In budding yeast, cyclin-CDK activity has been shown to have a crucial role in meiotic events such as formation of meiotic double-strand breaks that initiate homologous recombination. Mammalian cells express numerous cyclins and CDKs, but how these proteins control meiosis remains poorly understood. Cyclin B3 was previously identified as germ cell specific, and its restricted expression pattern at the beginning of meiosis made it an interesting candidate to regulate meiotic events. -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

Molecular Effects of Isoflavone Supplementation Human Intervention Studies and Quantitative Models for Risk Assessment
Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis committee Promotors Prof. Dr Pieter van ‘t Veer Professor of Nutritional Epidemiology Wageningen University Prof. Dr Evert G. Schouten Emeritus Professor of Epidemiology and Prevention Wageningen University Co-promotors Dr Anouk Geelen Assistant professor, Division of Human Nutrition Wageningen University Dr Lydia A. Afman Assistant professor, Division of Human Nutrition Wageningen University Other members Prof. Dr Jaap Keijer, Wageningen University Dr Hubert P.J.M. Noteborn, Netherlands Food en Consumer Product Safety Authority Prof. Dr Yvonne T. van der Schouw, UMC Utrecht Dr Wendy L. Hall, King’s College London This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 20 June 2014 at 13.30 p.m. in the Aula. Vera van der Velpen Molecular effects of isoflavone supplementation: Human intervention studies and quantitative models for risk assessment 154 pages PhD thesis, Wageningen University, Wageningen, NL (2014) With references, with summaries in Dutch and English ISBN: 978-94-6173-952-0 ABSTRact Background: Risk assessment can potentially be improved by closely linked experiments in the disciplines of epidemiology and toxicology. -

PPEF1 Monoclonal Antibody (M01), Clone 1F6-1A5
PPEF1 monoclonal antibody (M01), clone 1F6-1A5 Catalog # : H00005475-M01 規格 : [ 100 ug ] List All Specification Application Image Product Mouse monoclonal antibody raised against a full length recombinant Western Blot (Transfected lysate) Description: PPEF1. Immunogen: PPEF1 (AAH36026, 1 a.a. ~ 653 a.a) full-length recombinant protein with GST tag. MW of the GST tag alone is 26 KDa. Sequence: MGCSSSSTKTRRSDTSLRAALIIQNWYRGYKARLKARQHYALTIFQSIEY ADEQGQMQLSTFFSFMLENYTHIHKEELELRNQSLESEQDMRDRWDYV DSIDVPDSYNGPRLQFPLTCTDIDLLLEAFKEQQILHAHYVLEVLFETKKV enlarge LKQMPNFTHIQTSPSKEVTICGDLHGKLDDLFLIFYKNGLPSERNPYVFNG Western Blot (Recombinant DFVDRGKNSIEILMILCVSFLVYPNDLHLNRGNHEDFMMNLRYGFTKEILH protein) KYKLHGKRILQILEEFYAWLPIGTIVDNEILVIHGGISETTDLNLLHRVERNK MKSVLIPPTETNRDHDTDSKHNKVGVTFNAHGRIKTNGSPTEHLTEHEWE ELISA QIIDILWSDPRGKNGCFPNTCRGGGCYFGPDVTSKILNKYQLKMLIRSHE CKPEGYEICHDGKVVTIFSASNYYEEGSNRGAYIKLCSGTTPRFFQYQVT RNAi Knockdown (Antibody KATCFQPLRQRVDTMENSAIKILRERVISRKSDLTRAFQLQDHRKSGKLS validated) VSQWAFCMENILGLNLPWRSLSSNLVNIDQNGNVEYMSSFQNIRIEKPVQ EAHSTLVETLYRYRSDLEIIFNAIDTDHSGLISVEEFRAMWKLFSSHYNVHI DDSQVNKLANIMDLNKDGSIDFNEFLKAFYVVHRYEDLMKPDVTNLG Host: Mouse Reactivity: Human enlarge Isotype: IgG1 Kappa Quality Control Antibody Reactive Against Recombinant Protein. Testing: Western Blot detection against Immunogen (97.57 KDa) . Storage Buffer: In 1x PBS, pH 7.4 Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. Instruction: MSDS: Download Datasheet: Download Applications Page 1 of 3 2016/5/21 Western Blot (Transfected lysate) -

Frequent Loss of Heterozygosity Targeting the Inactive X Chromosome in Melanoma
6476 Vol. 9, 6476–6482, December 15, 2003 Clinical Cancer Research Frequent Loss of Heterozygosity Targeting the Inactive X Chromosome in Melanoma James O. Indsto,1 Najah T. Nassif,2 INTRODUCTION Richard F. Kefford,1 and Graham J. Mann1 Chromosomal deletions and amplification events are fre- 1Westmead Institute for Cancer Research, University of Sydney at quent in advanced neoplasms and provide evidence for the Westmead Millennium Institute, Westmead, New South Wales, existence and location of putative tumor suppressor genes Australia, and 2Department of Cell and Molecular Biology, University (TSGs) and oncogenes. In melanoma, 9p and 10q deletions are of Technology, Sydney, New South Wales, Australia most frequent, and losses on 6q, 11q, 1p, 15p, 17q, and 18q are also frequently found (1, 2). However, no studies have focused on the X chromosome. In the human female, random inactiva- ABSTRACT tion of one copy of an X chromosome by hypermethylation is an After previous preliminary observations of paradoxical early event in embryogenesis, resulting in most tissues being a deletion events affecting the inactive X chromosome in mel- mosaic in terms of X inactivation status. Neoplasms, being anoma, we have surveyed the X chromosome for deletions clonal expansions of single precursor cells, usually share a using 23 polymorphic microsatellite markers in 28 inform- common X inactivation status. This can be conveniently assayed ative (female XX) metastatic melanomas. Ten tumors (36%) using the methylation-sensitive restriction enzyme HpaII, which showed at least one loss of heterozygosity (LOH) event, and has a recognition site within the highly polymorphic exon 1 in two cases an entire chromosome showed LOH at all CAG repeat (ARTR) in the androgen receptor (AR) gene. -

75 2. INTRODUCTION Triple-Negative Breast Cancer (TNBC)
[Frontiers in Bioscience, Scholar, 11, 75-88, March 1, 2019] The persisting puzzle of racial disparity in triple negative breast cancer: looking through a new lens Chakravarthy Garlapati1, Shriya Joshi1, Bikram Sahoo1, Shobhna Kapoor2, Ritu Aneja1 1Department of Biology, Georgia State University, Atlanta, GA, USA, 2Department of Chemistry, Indian Institute of Technology Bombay, Powai, India TABLE OF CONTENTS 1. Abstract 2. Introduction 3. Dissecting the TNBC racially disparate burden 3.1. Does race influence TNBC onset and progression? 3.2. Tumor microenvironment in TNBC and racial disparity 3.3. Differential gene signatures and pathways in racially distinct TNBC 3.4. Our Perspective: Looking racial disparity through a new lens 4. Conclusion 5. Acknowledgement 6. References 1. ABSTRACT 2. INTRODUCTION Triple-negative breast cancer (TNBC) Triple-negative breast cancer (TNBC), is characterized by the absence of estrogen a subtype of breast cancer (BC), accounts for and progesterone receptors and absence 15-20% of all BC diagnoses in the US. It has of amplification of human epidermal growth been recognized that women of African descent factor receptor (HER2). This disease has no are twice as likely to develop TNBC than approved treatment with a poor prognosis women of European descent (1). As the name particularly in African-American (AA) as foretells, TNBCs lack estrogen, progesterone, compared to European-American (EA) and human epidermal growth factor receptors. patients. Gene ontology analysis showed Unfortunately, TNBCs are defined by what they specific gene pathways that are differentially “lack” rather than what they “have” and thus this regulated and gene signatures that are negative nomenclature provides no actionable differentially expressed in AA as compared to information on “druggable” targets.