Mouse Tubgcp3 Conditional Knockout Project (CRISPR/Cas9)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Subcellular Localization and Mitotic Interactome Analyses Identify SIRT4 As a Centrosomally Localized and Microtubule Associated Protein
cells Article Subcellular Localization and Mitotic Interactome Analyses Identify SIRT4 as a Centrosomally Localized and Microtubule Associated Protein 1 1, 1 1 Laura Bergmann , Alexander Lang y , Christoph Bross , Simone Altinoluk-Hambüchen , Iris Fey 1, Nina Overbeck 2, Anja Stefanski 2, Constanze Wiek 3, Andreas Kefalas 1, Patrick Verhülsdonk 1, Christian Mielke 4, Dennis Sohn 5, Kai Stühler 2,6, Helmut Hanenberg 3,7, Reiner U. Jänicke 5, Jürgen Scheller 1, Andreas S. Reichert 8 , Mohammad Reza Ahmadian 1 and Roland P. Piekorz 1,* 1 Institute of Biochemistry and Molecular Biology II, Medical Faculty, Heinrich Heine University Düsseldorf, 40225 Düsseldorf, Germany; [email protected] (L.B.); [email protected] (A.L.); [email protected] (C.B.); [email protected] (S.A.-H.); [email protected] (I.F.); [email protected] (A.K.); [email protected] (P.V.); [email protected] (J.S.); [email protected] (M.R.A.) 2 Molecular Proteomics Laboratory, Heinrich Heine University Düsseldorf, 40225 Düsseldorf, Germany; [email protected] (N.O.); [email protected] (A.S.); [email protected] (K.S.) 3 Department of Otolaryngology and Head/Neck Surgery, Medical Faculty, Heinrich Heine University Düsseldorf, 40225 Düsseldorf, Germany; [email protected] (C.W.); [email protected] (H.H.) 4 Institute of Clinical Chemistry and Laboratory Diagnostics, Medical Faculty, Heinrich Heine University Düsseldorf, 40225 Düsseldorf, Germany; [email protected] -

Iii COMPUTATIONAL APPROACHES for ASSESSING KINOME
COMPUTATIONAL APPROACHES FOR ASSESSING KINOME FUNCTION AND DEREGULATION A Dissertation Presented to the Faculty of the Weill Cornell Graduate School of Medical Sciences in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by Charles Joseph Murphy August 2018 iii © 2018 Charles Joseph Murphy ALL RIGHTS RESERVED COMPUTATIONAL APPROACHES FOR ASSESSING KINOME FUNCTION AND DEREGULATION Charles Joseph Murphy, PhD Cornell University 2018 Protein kinases are a diverse family of about 500 proteins that all share the common ability to catalyze phosphorylation of the side chains of amino acids in proteins. Kinases play a vital role across diverse biological functions including proliferation, differentiation, cell migration, and cell-cycle control. Moreover, they are frequently altered across most cancers types and have been a focus for development of anti- cancer drugs, which has led to the development of 38 approved kinase inhibitors as of 2018. In this thesis, I developed two orthogonal computational approaches for investigating kinase function and deregulation. Starting with data from a large cohort of mouse triple negative breast cancer (TNBC) tumors, I use a combination of whole exome sequencing (WES) and RNA-seq to identify somatic alterations that drive individual tumors. I discovered that a large number of these alterations involve protein kinases and subsequent therapeutic targeting led to tumor regression. For my second approach, I utilized a large peptide library dataset from about 300 kinases. Which kinase phosphorylate which phosphorylation site is determined by both kinase- intrinsic and contextual factors. Peptide library approaches provide kinase-intrinsic amino acid specificity, which I used to predict novel kinase substrates and map out kinase phosphorylation networks. -

Sirt1 Is Regulated by Mir-135A and Involved in DNA Damage Repair During Mouse Cellular Reprogramming
www.aging-us.com AGING 2020, Vol. 12, No. 8 Research Paper Sirt1 is regulated by miR-135a and involved in DNA damage repair during mouse cellular reprogramming Andy Chun Hang Chen1,2,*, Qian Peng2,*, Sze Wan Fong1, William Shu Biu Yeung1,2, Yin Lau Lee1,2 1Department of Obstetrics and Gynaecology, The University of Hong Kong, Hong Kong SAR, China 2Shenzhen Key Laboratory of Fertility Regulation, The University of Hong Kong Shenzhen Hospital, Shenzhen, China *Co-first authors Correspondence to: Yin Lau Lee, William Shu Biu Yeung; email: [email protected], [email protected] Keywords: mouse induced pluripotent stem cells, cellular reprogramming, Sirt1, miR-135a, DNA damage repair Received: December 30, 2019 Accepted: March 30, 2020 Published: April 26, 2020 Copyright: Chen et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Sirt1 facilitates the reprogramming of mouse somatic cells into induced pluripotent stem cells (iPSCs). It is regulated by micro-RNA and reported to be a target of miR-135a. However, their relationship and roles on cellular reprogramming remain unknown. In this study, we found negative correlations between miR-135a and Sirt1 during mouse embryonic stem cells differentiation and mouse embryonic fibroblasts reprogramming. We further found that the reprogramming efficiency was reduced by the overexpression of miR-135a precursor but induced by the miR-135a inhibitor. Co-immunoprecipitation followed by mass spectrometry identified 21 SIRT1 interacting proteins including KU70 and WRN, which were highly enriched for DNA damage repair. -
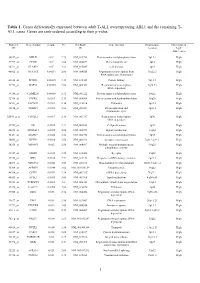
Cases. Genes Are Rank-Ordered According to Their P-Value
Table 1. Genes differentially expressed between adult T-ALL overexpressing ABL1 and the remaining T- ALL cases. Genes are rank-ordered according to their p-value. Probeset Gene symbol p-value FC GenBank Gene function Chromosome Expression in ID ID location “high” ABL1 cases 38847_at MELK <10-4 2.79 NM_014791 Protein amino acid phosphorylation 9p13.2 High 37999_at CPOX <10-4 2.86 NM_000097 Heme biosynthesis 3q12 High 38711_at CLASP2 <10-4 2.33 NM_015097 Cell division 3p23 High 40822_at NFATC3 0.00011 2.06 NM_004555 Regulation of transcription from 16q22.2 High RNA polymerase II promoter 41638_at PPWD1 0.00025 2.13 NM_015342 Protein folding 5q12.3 High 41278_at BAF53 0.00085 2.08 NM_004301 Regulation of transcription, 3q26.33 High DNA-dependent 39388_at CAMK2G 0.00088 2.32 NM_001222 Protein amino acid phosphorylation 10q22 High 32916_at PTPRE 0.0011 2.13 NM_006504 Protein amino acid dephosphorylation 10q26 High 36511_at SACM1L 0.0014 2.16 NM_014016 Unknown 3p21.3 High 38834_at TOPBP1 0.0015 2.55 NM_007027 DNA replication and 3q22.1 High chromosome cycle 33301_g_at CDC2L1 0.0017 2.16 NM_001787 Regulation of transcription, 1p36 High DNA-dependent 32767_at SIL 0.0025 2.23 NM_003035 Cell proliferation 1p32 High 40828_at ARHGEF7 0.0025 2.08 NM_003899 Signal transduction 13q34 High 38431_at MAPK9 0.0026 2.24 NM_002752 Protein amino acid phosphorylation 5q35 High 35663_at NPTX2 0.0028 3.68 NM_002523 Synaptic transmission 7q21.3-q22.1 High 38325_at MINPP1 0.003 2.58 NM_004897 Multiple inositol-polyphosphate 10q23 High phosphatase activity 35747_at -

UC San Diego UC San Diego Electronic Theses and Dissertations
UC San Diego UC San Diego Electronic Theses and Dissertations Title Regulation of gene expression programs by serum response factor and megakaryoblastic leukemia 1/2 in macrophages Permalink https://escholarship.org/uc/item/8cc7d0t0 Author Sullivan, Amy Lynn Publication Date 2009 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO Regulation of Gene Expression Programs by Serum Response Factor and Megakaryoblastic Leukemia 1/2 in Macrophages A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Biomedical Sciences by Amy Lynn Sullivan Committee in charge: Professor Christopher K. Glass, Chair Professor Stephen M. Hedrick Professor Marc R. Montminy Professor Nicholas J. Webster Professor Joseph L. Witztum 2009 Copyright Amy Lynn Sullivan, 2009 All rights reserved. The Dissertation of Amy Lynn Sullivan is approved, and it is acceptable in quality and form for publication on microfilm and electronically: ______________________________________________________________ ______________________________________________________________ ______________________________________________________________ ______________________________________________________________ ______________________________________________________________ Chair University of California, San Diego 2009 iii DEDICATION To my husband, Shane, for putting up with me through all of the long hours, last minute late nights, and for not letting me quit no matter how many times my projects fell apart. To my son, Tyler, for always making me smile and for making every day an adventure. To my gifted colleagues, for all of the thought-provoking discussions, technical help and moral support through the roller- coaster ride that has been my graduate career. To my family and friends, for all of your love and support. I couldn’t have done it without you! iv EPIGRAPH If at first you don’t succeed, try, try, again. -

Suppl. Table 1
Suppl. Table 1. SiRNA library used for centriole overduplication screen. Entrez Gene Id NCBI gene symbol siRNA Target Sequence 1070 CETN3 TTGCGACGTGTTGCTAGAGAA 1070 CETN3 AAGCAATAGATTATCATGAAT 55722 CEP72 AGAGCTATGTATGATAATTAA 55722 CEP72 CTGGATGATTTGAGACAACAT 80071 CCDC15 ACCGAGTAAATCAACAAATTA 80071 CCDC15 CAGCAGAGTTCAGAAAGTAAA 9702 CEP57 TAGACTTATCTTTGAAGATAA 9702 CEP57 TAGAGAAACAATTGAATATAA 282809 WDR51B AAGGACTAATTTAAATTACTA 282809 WDR51B AAGATCCTGGATACAAATTAA 55142 CEP27 CAGCAGATCACAAATATTCAA 55142 CEP27 AAGCTGTTTATCACAGATATA 85378 TUBGCP6 ACGAGACTACTTCCTTAACAA 85378 TUBGCP6 CACCCACGGACACGTATCCAA 54930 C14orf94 CAGCGGCTGCTTGTAACTGAA 54930 C14orf94 AAGGGAGTGTGGAAATGCTTA 5048 PAFAH1B1 CCCGGTAATATCACTCGTTAA 5048 PAFAH1B1 CTCATAGATATTGAACAATAA 2802 GOLGA3 CTGGCCGATTACAGAACTGAA 2802 GOLGA3 CAGAGTTACTTCAGTGCATAA 9662 CEP135 AAGAATTTCATTCTCACTTAA 9662 CEP135 CAGCAGAAAGAGATAAACTAA 153241 CCDC100 ATGCAAGAAGATATATTTGAA 153241 CCDC100 CTGCGGTAATTTCCAGTTCTA 80184 CEP290 CCGGAAGAAATGAAGAATTAA 80184 CEP290 AAGGAAATCAATAAACTTGAA 22852 ANKRD26 CAGAAGTATGTTGATCCTTTA 22852 ANKRD26 ATGGATGATGTTGATGACTTA 10540 DCTN2 CACCAGCTATATGAAACTATA 10540 DCTN2 AACGAGATTGCCAAGCATAAA 25886 WDR51A AAGTGATGGTTTGGAAGAGTA 25886 WDR51A CCAGTGATGACAAGACTGTTA 55835 CENPJ CTCAAGTTAAACATAAGTCAA 55835 CENPJ CACAGTCAGATAAATCTGAAA 84902 CCDC123 AAGGATGGAGTGCTTAATAAA 84902 CCDC123 ACCCTGGTTGTTGGATATAAA 79598 LRRIQ2 CACAAGAGAATTCTAAATTAA 79598 LRRIQ2 AAGGATAATATCGTTTAACAA 51143 DYNC1LI1 TTGGATTTGTCTATACATATA 51143 DYNC1LI1 TAGACTTAGTATATAAATACA 2302 FOXJ1 CAGGACAGACAGACTAATGTA -

Peripheral Blood Epi-Signature of Claes-Jensen Syndrome Enables Sensitive and Specific Identification of Patients and Healthy Ca
Schenkel et al. Clinical Epigenetics (2018) 10:21 https://doi.org/10.1186/s13148-018-0453-8 RESEARCH Open Access Peripheral blood epi-signature of Claes- Jensen syndrome enables sensitive and specific identification of patients and healthy carriers with pathogenic mutations in KDM5C Laila C. Schenkel1,2†, Erfan Aref-Eshghi1,2†, Cindy Skinner3, Peter Ainsworth1,2, Hanxin Lin1,2, Guillaume Paré4, David I. Rodenhiser5, Charles Schwartz3 and Bekim Sadikovic1,2,6* Abstract Background: Claes-Jensen syndrome is an X-linked inherited intellectual disability caused by mutations in the KDM5C gene. Kdm5c is a histone lysine demethylase involved in histone modifications and chromatin remodeling. Males with hemizygous mutations in KDM5C present with intellectual disability and facial dysmorphism, while most heterozygous female carriers are asymptomatic. We hypothesized that loss of Kdm5c function may influence other components of the epigenomic machinery including DNA methylation in affected patients. Results: Genome-wide DNA methylation analysis of 7 male patients affected with Claes-Jensen syndrome and 56 age- and sex-matched controls identified a specific DNA methylation defect (epi-signature) in the peripheral blood of these patients, including 1769 individual CpGs and 9 genomic regions. Six healthy female carriers showed less pronounced but distinctive changes in the same regions enabling their differentiation from both patients and controls. Highly specific computational model using the most significant methylation changes demonstrated 100% accuracy in differentiating patients, carriers, and controls in the training cohort, which was confirmed on a separate cohort of patients and carriers. The 100% specificity of this unique epi-signature was further confirmed on additional 500 unaffected controls and 600 patients with intellectual disability and developmental delay, including other patient cohorts with previously described epi-signatures. -

Novel Targets of Apparently Idiopathic Male Infertility
International Journal of Molecular Sciences Review Molecular Biology of Spermatogenesis: Novel Targets of Apparently Idiopathic Male Infertility Rossella Cannarella * , Rosita A. Condorelli , Laura M. Mongioì, Sandro La Vignera * and Aldo E. Calogero Department of Clinical and Experimental Medicine, University of Catania, 95123 Catania, Italy; [email protected] (R.A.C.); [email protected] (L.M.M.); [email protected] (A.E.C.) * Correspondence: [email protected] (R.C.); [email protected] (S.L.V.) Received: 8 February 2020; Accepted: 2 March 2020; Published: 3 March 2020 Abstract: Male infertility affects half of infertile couples and, currently, a relevant percentage of cases of male infertility is considered as idiopathic. Although the male contribution to human fertilization has traditionally been restricted to sperm DNA, current evidence suggest that a relevant number of sperm transcripts and proteins are involved in acrosome reactions, sperm-oocyte fusion and, once released into the oocyte, embryo growth and development. The aim of this review is to provide updated and comprehensive insight into the molecular biology of spermatogenesis, including evidence on spermatogenetic failure and underlining the role of the sperm-carried molecular factors involved in oocyte fertilization and embryo growth. This represents the first step in the identification of new possible diagnostic and, possibly, therapeutic markers in the field of apparently idiopathic male infertility. Keywords: spermatogenetic failure; embryo growth; male infertility; spermatogenesis; recurrent pregnancy loss; sperm proteome; DNA fragmentation; sperm transcriptome 1. Introduction Infertility is a widespread condition in industrialized countries, affecting up to 15% of couples of childbearing age [1]. It is defined as the inability to achieve conception after 1–2 years of unprotected sexual intercourse [2]. -

Anti-TUBGCP4 Monoclonal Antibody, Clone HU923 (DCABH-7801) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Anti-TUBGCP4 monoclonal antibody, clone HU923 (DCABH-7801) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Product Overview Mouse monoclonal to GCP4 Antigen Description GCP4 is a component of the gamma-tubulin complex which is necessary for microtubule nucleation at the centrosome. GCP4 shows sequence identity with the C-terminal regions of human centrosomal proteins 103p (TUBGCP2) and 104p (TUBGCP3). Immunogen Recombinant fragment within Human GCP4 aa 1-178. The exact sequence is proprietary.Database link: Q9UGJ1 Isotype IgG2b Source/Host Mouse Species Reactivity Human Clone HU923 Purity Protein G purified Conjugate Unconjugated Applications WB, ICC/IF Positive Control HeLa cell lysate; HeLa cells. Format Liquid Size 100 μl Preservative None Storage Store at +4°C short term (1-2 weeks). Upon delivery aliquot. Store at -20°C long term. Avoid freeze / thaw cycle. Ship Shipped at 4°C. GENE INFORMATION 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved Gene Name TUBGCP4 tubulin, gamma complex associated protein 4 [ Homo sapiens ] Official Symbol TUBGCP4 Synonyms TUBGCP4; tubulin, gamma complex associated protein 4; gamma-tubulin complex component 4; 76P; FLJ14797; gamma-ring complex protein 76 kDa; gamma tubulin ring complex protein (76p gene); GCP4; h76p; GCP-4; hGCP4; hGrip76; Entrez Gene ID 27229 Protein Refseq NP_055259 UniProt ID Q9UGJ1 Chromosome Location 15q15 Pathway Cell Cycle, organism-specific biosystem; Cell Cycle, Mitotic, organism-specific biosystem; Centrosome maturation, organism-specific biosystem; G2/M Transition, organism-specific biosystem; Mitotic G2-G2/M phases, organism-specific biosystem; Recruitment of NuMA to mitotic centrosomes, organism-specific biosystem; Recruitment of mitotic centrosome proteins and complexes, organism-specific biosystem. -

Molecular Targeting and Enhancing Anticancer Efficacy of Oncolytic HSV-1 to Midkine Expressing Tumors
University of Cincinnati Date: 12/20/2010 I, Arturo R Maldonado , hereby submit this original work as part of the requirements for the degree of Doctor of Philosophy in Developmental Biology. It is entitled: Molecular Targeting and Enhancing Anticancer Efficacy of Oncolytic HSV-1 to Midkine Expressing Tumors Student's name: Arturo R Maldonado This work and its defense approved by: Committee chair: Jeffrey Whitsett Committee member: Timothy Crombleholme, MD Committee member: Dan Wiginton, PhD Committee member: Rhonda Cardin, PhD Committee member: Tim Cripe 1297 Last Printed:1/11/2011 Document Of Defense Form Molecular Targeting and Enhancing Anticancer Efficacy of Oncolytic HSV-1 to Midkine Expressing Tumors A dissertation submitted to the Graduate School of the University of Cincinnati College of Medicine in partial fulfillment of the requirements for the degree of DOCTORATE OF PHILOSOPHY (PH.D.) in the Division of Molecular & Developmental Biology 2010 By Arturo Rafael Maldonado B.A., University of Miami, Coral Gables, Florida June 1993 M.D., New Jersey Medical School, Newark, New Jersey June 1999 Committee Chair: Jeffrey A. Whitsett, M.D. Advisor: Timothy M. Crombleholme, M.D. Timothy P. Cripe, M.D. Ph.D. Dan Wiginton, Ph.D. Rhonda D. Cardin, Ph.D. ABSTRACT Since 1999, cancer has surpassed heart disease as the number one cause of death in the US for people under the age of 85. Malignant Peripheral Nerve Sheath Tumor (MPNST), a common malignancy in patients with Neurofibromatosis, and colorectal cancer are midkine- producing tumors with high mortality rates. In vitro and preclinical xenograft models of MPNST were utilized in this dissertation to study the role of midkine (MDK), a tumor-specific gene over- expressed in these tumors and to test the efficacy of a MDK-transcriptionally targeted oncolytic HSV-1 (oHSV). -

Supplemental Solier
Supplementary Figure 1. Importance of Exon numbers for transcript downregulation by CPT Numbers of down-regulated genes for four groups of comparable size genes, differing only by the number of exons. Supplementary Figure 2. CPT up-regulates the p53 signaling pathway genes A, List of the GO categories for the up-regulated genes in CPT-treated HCT116 cells (p<0.05). In bold: GO category also present for the genes that are up-regulated in CPT- treated MCF7 cells. B, List of the up-regulated genes in both CPT-treated HCT116 cells and CPT-treated MCF7 cells (CPT 4 h). C, RT-PCR showing the effect of CPT on JUN and H2AFJ transcripts. Control cells were exposed to DMSO. β2 microglobulin (β2) mRNA was used as control. Supplementary Figure 3. Down-regulation of RNA degradation-related genes after CPT treatment A, “RNA degradation” pathway from KEGG. The genes with “red stars” were down- regulated genes after CPT treatment. B, Affy Exon array data for the “CNOT” genes. The log2 difference for the “CNOT” genes expression depending on CPT treatment was normalized to the untreated controls. C, RT-PCR showing the effect of CPT on “CNOT” genes down-regulation. HCT116 cells were treated with CPT (10 µM, 20 h) and CNOT6L, CNOT2, CNOT4 and CNOT6 mRNA were analysed by RT-PCR. Control cells were exposed to DMSO. β2 microglobulin (β2) mRNA was used as control. D, CNOT6L down-regulation after CPT treatment. CNOT6L transcript was analysed by Q- PCR. Supplementary Figure 4. Down-regulation of ubiquitin-related genes after CPT treatment A, “Ubiquitin-mediated proteolysis” pathway from KEGG. -

Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared Amongst Δγ T, Innate Lymphoid, and Th Cells
Downloaded from http://www.jimmunol.org/ by guest on September 26, 2021 δγ is online at: average * The Journal of Immunology , 10 of which you can access for free at: 2016; 197:1460-1470; Prepublished online 6 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600643 http://www.jimmunol.org/content/197/4/1460 Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst T, Innate Lymphoid, and Th Cells You Jeong Lee, Gabriel J. Starrett, Seungeun Thera Lee, Rendong Yang, Christine M. Henzler, Stephen C. Jameson and Kristin A. Hogquist J Immunol cites 41 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2016/07/06/jimmunol.160064 3.DCSupplemental This article http://www.jimmunol.org/content/197/4/1460.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 26, 2021. The Journal of Immunology Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst gd T, Innate Lymphoid, and Th Cells You Jeong Lee,* Gabriel J.