Comprehensive Testing for NF1/SPRED1 and Other Rasopathies Genes at UAB
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
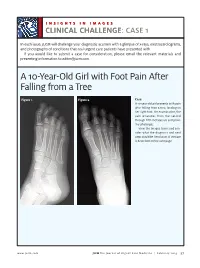
A 10-Year-Old Girl with Foot Pain After Falling from a Tree
INSIGHTS IN IMAGES CLINICAL CHALLENGECHALLENGE: CASE 1 In each issue, JUCM will challenge your diagnostic acumen with a glimpse of x-rays, electrocardiograms, and photographs of conditions that real urgent care patients have presented with. If you would like to submit a case for consideration, please email the relevant materials and presenting information to [email protected]. A 10-Year-Old Girl with Foot Pain After Falling from a Tree Figure 1. Figure 2. Case A 10-year-old girl presents with pain after falling from a tree, landing on her right foot. On examination, the pain emanates from the second through fifth metatarsals and proxi- mal phalanges. View the images taken and con- sider what the diagnosis and next steps would be. Resolution of the case is described on the next page. www.jucm.com JUCM The Journal of Urgent Care Medicine | February 2019 37 INSIGHTS IN IMAGES: CLINICAL CHALLENGE THE RESOLUTION Figure 1. Mediastinal air Figure 1. Differential Diagnosis Pearls for Urgent Care Management and Ⅲ Fracture of the distal fourth metatarsal Considerations for Transfer Ⅲ Plantar plate disruption Ⅲ Emergent transfer should be considered with associated neu- Ⅲ Sesamoiditis rologic deficit, compartment syndrome, open fracture, or vas- Ⅲ Turf toe cular compromise Ⅲ Referral to an orthopedist is warranted in the case of an in- Diagnosis tra-articular fracture, or with Lisfranc ligament injury or ten- Angulation of the distal fourth metatarsal metaphyseal cortex derness over the Lisfranc ligament and hairline lucency consistent with fracture. Acknowledgment: Images courtesy of Teleradiology Associates. Learnings/What to Look for Ⅲ Proximal metatarsal fractures are most often caused by crush- ing or direct blows Ⅲ In athletes, an axial load placed on a plantar-flexed foot should raise suspicion of a Lisfranc injury 38 JUCM The Journal of Urgent Care Medicine | February 2019 www.jucm.com INSIGHTS IN IMAGES CLINICAL CHALLENGE: CASE 2 A 55-Year-Old Man with 3 Hours of Epigastric Pain 55 years PR 249 QRSD 90 QT 471 QTc 425 AXES P 64 QRS -35 T 30 Figure 1. -

Genetic Mechanisms of Disease in Children: a New Look
Genetic Mechanisms of Disease in Children: A New Look Laurie Demmer, MD Tufts Medical Center and the Floating Hospital for Children Boston, MA Traditionally genetic disorders have been linked to the ‘one gene-one protein-one disease’ hypothesis. However recent advances in the field of molecular biology and biotechnology have afforded us the opportunity to greatly expand our knowledge of genetics, and we now know that the mechanisms of inherited disorders are often significantly more complex, and consequently, much more intriguing, than originally thought. Classical mendelian disorders with relatively simple genetic mechanisms do exist, but turn out to be far more rare than originally thought. All patients with sickle cell disease for example, carry the same A-to-T point mutation in the sixth codon of the beta globin gene. This results in a glutamate to valine substitution which changes the shape and the function of the globin molecule in a predictable way. Similarly, all patients with achondroplasia have a single base pair substitution at nucleotide #1138 of the FGFR3 gene. On the other hand, another common inherited disorder, cystic fibrosis, is known to result from changes in a specific transmembrane receptor (CFTR), but over 1000 different disease-causing mutations have been reported in this single gene. Since most commercial labs only test for between 23-100 different mutations, interpreting CFTR mutation testing is significantly complicated by the known risk of false negative results. Many examples of complex, or non-Mendelian, inheritance are now known to exist and include disorders of trinucleotide repeats, errors in imprinting, and gene dosage effects. -

Segmental Overvekst Og Vaskulærmalformasjoner V02
2/1/2021 Segmental overvekst og vaskulærmalformasjoner v02 Avdeling for medisinsk genetikk Segmental overvekst og vaskulærmalformasjoner Genpanel, versjon v02 * Enkelte genomiske regioner har lav eller ingen sekvensdekning ved eksomsekvensering. Dette skyldes at de har stor likhet med andre områder i genomet, slik at spesifikk gjenkjennelse av disse områdene og påvisning av varianter i disse områdene, blir vanskelig og upålitelig. Disse genetiske regionene har vi identifisert ved å benytte USCS segmental duplication hvor områder større enn 1 kb og ≥90% likhet med andre regioner i genomet, gjenkjennes (https://genome.ucsc.edu). Vi gjør oppmerksom på at ved identifiseringav ekson oppstrøms for startkodon kan eksonnummereringen endres uten at transkript ID endres. Avdelingens websider har en full oversikt over områder som er affisert av segmentale duplikasjoner. ** Transkriptets kodende ekson. Ekson Gen Gen affisert (HGNC (HGNC Transkript Ekson** Fenotype av symbol) ID) segdup* ACVRL1 175 NM_000020.3 2-10 Telangiectasia, hereditary hemorrhagic, type 2 OMIM ADAMTS3 219 NM_014243.3 1-22 Hennekam lymphangiectasia- lymphedema syndrome 3 OMIM AKT1 391 NM_005163.2 2-14 Cowden syndrome 6 OMIM Proteus syndrome, somatic OMIM AKT2 392 NM_001626.6 2-14 Diabetes mellitus, type II OMIM Hypoinsulinemic hypoglycemia with hemihypertrophy OMIM AKT3 393 NM_005465.7 2-14 Megalencephaly-polymicrogyria- polydactyly-hydrocephalus syndrome 2 OMIM file:///data/SegOv_v02-web.html 1/7 2/1/2021 Segmental overvekst og vaskulærmalformasjoner v02 Ekson Gen Gen affisert (HGNC (HGNC -

Master List of References ACADVL 1
Last Revision: 6/2017 DEPARTMENT OF PATHOLOGY AND LABORATORY MEDICINE PRECISION DIAGNOSTICS – INHERITED DISEASE Master List of References ACADVL 1. Mathur A, Sims HF, Gopalakrishnan D et al. Molecular heterogeneity in very-long-chain acyl-CoA dehydrogenase deficiency causing pediatric cardiomyopathy and sudden death. Circulation 1999;99:1337-43. [PMID: 10077518] 2. Vianey-Saban C, Divry P, Brivet M et al. Mitochondrial very-long-chain acyl-coenzyme A dehydrogenase deficiency: Clinical characteristics and diagnostic considerations in 30 patients. Clin Chim Acta 1998;269:43-62. [PMID: 9498103] AR 1. Giagulli VA, Carbone MD, De Pergola G, et al. Could androgen receptor gene CAG tract polymorphism affect spermatogenesis in men with idiopathic infertility? J Assist Reprod Genet 2014;31(6):689-97. [PMID: 24691874] 2. Gottlieb B, Beitel LK, Nadarajah A, et al. The androgen receptor gene mutations database (ARDB): 2012 update. Hum Mutat. 2012;33:887-94. [PMID: 22334387] Array CGH 1. Boone PM, Bacino CA, Shaw CA et al., Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat 2010;31(12):1326-1342. [PMID: 20848651] CFTR 1. Collaco AM, Geibel P, Lee BS et al. Functional vacuolar ATPase (V-ATPase) proton pumps traffic to the enterocyte brush border membrane and require CFTR. Am J Physiol Cell Physiol 2013;305(9):C981-996. [PMID: 23986201] 2. Lu S, Yang X, Li X et al. Different cystic fibrosis transmembrane coductance regulator mutations in Chinese men with congenital bilateral absence of vas deferens and other acquired obtructive azoospermia. Urology 2013;82(4):824-828. [PMID: 23953609] 3. Sosnay PR, Siklosi KR, Van Goor F et al. -

Piggybac Mutagenesis and Exome Sequencing Identify Genetic Driver
Noorani et al. Genome Biology (2020) 21:181 https://doi.org/10.1186/s13059-020-02092-2 RESEARCH Open Access PiggyBac mutagenesis and exome sequencing identify genetic driver landscapes and potential therapeutic targets of EGFR-mutant gliomas Imran Noorani1,2* , Jorge de la Rosa1†, Yoon Ha Choi1,3†, Alexander Strong1, Hannes Ponstingl1, M. S. Vijayabaskar1, Jusung Lee3, Eunmin Lee3, Angela Richard-Londt4, Mathias Friedrich1,5, Federica Furlanetto5, Rocio Fuente1, Ruby Banerjee1, Fengtang Yang1, Frances Law1, Colin Watts2,6, Roland Rad5, George Vassiliou1, Jong Kyoung Kim3, Thomas Santarius2, Sebastian Brandner4 and Allan Bradley1* * Correspondence: [email protected]; [email protected] Abstract †Jorge de la Rosa and Yoonha Choi contributed equally to this work. Background: Glioma is the most common intrinsic brain tumor and also occurs in 1The Wellcome Trust Sanger the spinal cord. Activating EGFR mutations are common in IDH1 wild-type gliomas. Institute, Wellcome Trust Genome However, the cooperative partners of EGFR driving gliomagenesis remain poorly Campus, Hinxton, Cambridgeshire CB10 1SA, UK understood. Full list of author information is Results: We explore EGFR-mutant glioma evolution in conditional mutant mice by available at the end of the article whole-exome sequencing, transposon mutagenesis forward genetic screening, and transcriptomics. We show mutant EGFR is sufficient to initiate gliomagenesis in vivo, both in the brain and spinal cord. We identify significantly recurrent somatic alterations in these gliomas including mutant EGFR amplifications and Sub1, Trp53, and Tead2 loss-of-function mutations. Comprehensive functional characterization of 96 gliomas by genome-wide piggyBac insertional mutagenesis in vivo identifies 281 known and novel EGFR-cooperating driver genes, including Cdkn2a, Nf1, Spred1, and Nav3. -

Megalencephaly and Macrocephaly
277 Megalencephaly and Macrocephaly KellenD.Winden,MD,PhD1 Christopher J. Yuskaitis, MD, PhD1 Annapurna Poduri, MD, MPH2 1 Department of Neurology, Boston Children’s Hospital, Boston, Address for correspondence Annapurna Poduri, Epilepsy Genetics Massachusetts Program, Division of Epilepsy and Clinical Electrophysiology, 2 Epilepsy Genetics Program, Division of Epilepsy and Clinical Department of Neurology, Fegan 9, Boston Children’s Hospital, 300 Electrophysiology, Department of Neurology, Boston Children’s Longwood Avenue, Boston, MA 02115 Hospital, Boston, Massachusetts (e-mail: [email protected]). Semin Neurol 2015;35:277–287. Abstract Megalencephaly is a developmental disorder characterized by brain overgrowth secondary to increased size and/or numbers of neurons and glia. These disorders can be divided into metabolic and developmental categories based on their molecular etiologies. Metabolic megalencephalies are mostly caused by genetic defects in cellular metabolism, whereas developmental megalencephalies have recently been shown to be caused by alterations in signaling pathways that regulate neuronal replication, growth, and migration. These disorders often lead to epilepsy, developmental disabilities, and Keywords behavioral problems; specific disorders have associations with overgrowth or abnor- ► megalencephaly malities in other tissues. The molecular underpinnings of many of these disorders are ► hemimegalencephaly now understood, providing insight into how dysregulation of critical pathways leads to ► -

International Symposium on New Developments in Neurofibromatoses and Rasopathies
INTERNATIONAL SYMPOSIUM ON NEW DEVELOPMENTS IN NEUROFIBROMATOSES AND RASOPATHIES Their Management, Diagnosis, Current and Future Therapeutic Targets 27th - 29th November 2017, Crowne Plaza, Kochi, Kerala, South India INTERNATIONAL SYMPOSIUM ON NEW DEVELOPMENTS IN NEUROFIBROMATOSES AND RASOPATHIES THE FIRST INTERNATIONAL SYMPOSIUM ON RASOPATHIES TO BE HELD IN ASIA RASopathies are a class of developmental disorders with overlapping clinical features as well as genetic mutations. RASopathies are associated with dysregulation of the RAS-MAPK (RAS/mitogen activated protein kinase) signalling pathway, an important pathway in humans as it is related to nine different genetic conditions and in many cancers. The meeting will bring together leading scientists, clinicians, researchers and trainee health care professionals in the field of genetics, cancer, neurology, paediatrics, cardiology, neurosurgery, craniofacial surgery, plastic surgery, dermatology, oncology and biomedical sciences from across the world. 200 25+ 20+ PLACES SPEAKERS TOPICS AIMS TO ENSURE THAT HEALTH PROFESSIONALS AND TO IDENTIFY THERAPEUTIC TARGETS FOR SCIENTISTS ARE KEPT ABREAST OF ANY NEW SEVERAL OF THESE RARE DISEASES DEVELOPMENTS IN CLINICAL PRACTICE TOPICS COVERED o RASopathies: Syndromes of Ras/ MAPK pathway o Cardio-Facio-Cutaneous Syndrome dysregulation o Capillary Malformation-Arteriovenous o Neurofibromatosis Type 1 Malformation Syndrome o Legius Syndrome o Natural history, improved diagnosis and genotype phenotype correlation for NF2 and o Noonan Syndrome Schwannomatosis -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Essential Role of Autophagy in Protecting Neonatal Haematopoietic Stem Cells from Oxidative Stress in a P62-Independent Manner
www.nature.com/scientificreports OPEN Essential role of autophagy in protecting neonatal haematopoietic stem cells from oxidative stress in a p62‑independent manner Naho Nomura1,7,10, Chiaki Ito1,10, Takako Ooshio1,8, Yuko Tadokoro1,2, Susumu Kohno3, Masaya Ueno1,2, Masahiko Kobayashi1,2, Atsuko Kasahara4, Yusuke Takase1,9, Kenta Kurayoshi1, Sha Si1,2, Chiaki Takahashi3, Masaaki Komatsu5, Toru Yanagawa6 & Atsushi Hirao1,2* Autophagy is a cellular degradation system contributing to homeostasis of tissue stem cells including haematopoietic stem cells (HSCs). It plays pleiotropic roles in HSC characteristics throughout life, but its stage‑specifc roles in HSC self‑renewal are unclear. To investigate the efects of Atg5 deletion on stage‑specifc HSC functions, we compared the repopulating capacity of HSCs in Atg5f/f;Vavi-cre mice from postnatal day (P) 0–7 weeks of age. Interestingly, Atg5 defciency led to no remarkable abnormality in the HSC self‑renewal capacity at P0, but signifcant defects at P7, followed by severe defects. Induction of Atg5 deletion at P5 by tamoxifen administration to Atg5f/f;Rosa26-Cre-ERT2 mice resulted in normal haematopoiesis, including the HSC population, until around 1 year, suggesting that Atg5 in the early neonatal period was critical for haematopoiesis in adults. Mitochondrial oxidative stress was increased by Atg5 loss in neonatal HSC/progenitor cells. Although p62 had accumulated in immature bone marrow cells of Atg5f/f;Vavi-cre mice, p62 deletion did not restore defective HSC functions, indicating that Atg5‑dependent haematopoietic regulation in the developmental period was independent of p62. This study proposes a critical role of autophagy in HSC protection against harsh environments in the early neonatal stage, which is essential for healthy long‑term haematopoiesis. -

An Unusual Case of Cowden Syndrome Associated With
Pistorius et al. Hereditary Cancer in Clinical Practice (2016) 14:11 DOI 10.1186/s13053-016-0051-8 CASE REPORT Open Access An unusual case of Cowden syndrome associated with ganglioneuromatous polyposis Steffen Pistorius1,6*†, Barbara Klink2,7*†, Jessica Pablik3, Andreas Rump2, Daniela Aust3,7, Marlene Garzarolli4, Evelin Schröck2,7 and Hans K. Schackert5,6,7 Abstract Background: Ganglioneuromatous polyposis (GP) is a very rare disorder which may be associated with other clinical manifestations and syndromes, such as Cowden syndrome, multiple endocrine neoplasia (MEN) type II and neurofibromatosis (NF) 1. The risk for malignant transformation of ganglioneuromas is unknown, and the combination of GP with colon cancer has been only very seldom reported. Methods and results: We report the case of a 60-year old male patient with adenocarcinoma, adenomas and lipomas of the colon and multiple gastroduodenal lesions combined with generalised lipomatosis and macrocephaly. Based on the initial endoscopic and histological findings, a (restorative) proctocolectomy was recommended but declined by the patient. Instead, a colectomy was performed. The histological examination revealed an unforeseen GP in addition to the colon cancer. Extensive molecular diagnostics allowed for the differential diagnosis of the causes of the clinical manifestations, and the clinical suspicion of Cowden syndrome could not be confirmed using Sanger Sequencing and MLPA for the analysis of PTEN. Finally, a pathogenic germline mutation in PTEN (heterozygous stop mutation in exon 2: NM_000314 (PTEN):c.138C > A; p.Tyr46*) could be detected by next-generation sequencing (NGS), confirming an unusual presentation of Cowden syndrome with GP. Conclusions: Cowden syndrome should be considered in cases of GP with extracolonic manifestation and verified by combined clinical and molecular diagnostics. -

Costello Syndrome
orphananesthesia Anaesthesia recommendations for patients suffering from Costello syndrome Disease name: Costello syndrome ICD 10: Q87.8 Synonyms: Significant phenotypical overlap with CFC (cardiofaciocutaneous syndrome) and Noonan syndrome. Disease summary: Costello syndrome (CS) is a rare disorder (so-called RAS-opathy, see below), affecting up to 300 people worldwide. First described by Dr Jack Costello in 1977, the syndrome is characterised by failure to thrive (FTT), poor feeding, short stature, developmental delay, distinctive facial features, excessive loose skin, cardiac abnormalities, and an increased risk of tumour development. RAS is a family of genes coding for small GTPases and includes amongst others HRAS. The HRAS gene is a proto-oncogene, which forms part of the MAPK (mitogen activated protein kinase) signalling pathway. Up-regulation of this signalling pathway causes unopposed cell growth, causing tumour predisposition. The MAPK pathway is also the site of mutations causing both CFC and Noonan syndrome. CS can be caused by a number of mutations in the HRAS gene. Most mutations do occur de novo, but there is some evidence that a minority are inherited in an autosomal dominant manner. Patients with CS are born large for gestational age, and there is a strong association with polyhydramnios and preterm labour. Growth later slows due to feeding difficulties. Head circumference is affected to a lesser degree than height and weight, which gives rise to relative macrocephaly. Growth Hormone (GH) deficiency can cause neonatal hypoglycaemia, and contributes to growth retardation. The disease is characterised by distinctive facial features including downslanting palpebral fissures, epicanthic folds, ptosis, flattened nasal bridge (hypertelorism), low set ears, thick lips, macroglossia and short neck. -

Mir-126-3P Contributes to Sorafenib Resistance in Hepatocellular Carcinoma Via Downregulating SPRED1
38 Original Article Page 1 of 14 miR-126-3p contributes to sorafenib resistance in hepatocellular carcinoma via downregulating SPRED1 Wenliang Tan1,2#, Zhirong Lin1,2#, Xianqing Chen3, Wenxin Li4, Sicong Zhu1,5, Yingcheng Wei1,2, Liyun Huo1,2, Yajin Chen1,2, Changzhen Shang1,2 1Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-sen Memorial Hospital, Sun Yat-Sen University, Guangzhou, China; 2Department of Hepatobiliary Surgery, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China; 3Department of Hepatobiliary Surgery, the Eighth Affiliated Hospital, Sun Yat-sen University, Shenzhen, China; 4Department of Cardiology, the Eighth Affiliated Hospital, Sun Yat-sen University, Shenzhen, China; 5Department of Surgical Intensive Care Unit, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China Contributions: (I) Conception and design: C Shang, Y Chen; (II) Administrative support: C Shang, Y Chen; (III) Provision of study materials or patients: W Tan, Z Lin; (IV) Collection and assembly of data: S Zhu, Y Wei, L Huo; (V) Data analysis and interpretation: X Chen, W Li; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. #These authors contributed equally to this work. Correspondence to: Changzhen Shang; Yajin Chen. Department of Hepatobiliary Surgery, Sun Yat-sen Memorial Hospital of Sun Yat-sen University, Guangzhou 510120, China. Email: [email protected]; [email protected]. Background: Sorafenib can prolong the survival of patients with advanced hepatocellular carcinoma (HCC). However, drug resistance remains the main obstacle to improving its efficiency. This study aimed to explore the likely molecular mechanism of sorafenib resistance. Methods: Differentially expressed microRNAs (miRNAs) related to sorafenib response were analyzed with the Limma package in R software.