Segmental Overvekst Og Vaskulærmalformasjoner V02
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
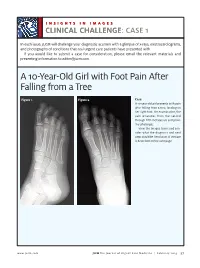
A 10-Year-Old Girl with Foot Pain After Falling from a Tree
INSIGHTS IN IMAGES CLINICAL CHALLENGECHALLENGE: CASE 1 In each issue, JUCM will challenge your diagnostic acumen with a glimpse of x-rays, electrocardiograms, and photographs of conditions that real urgent care patients have presented with. If you would like to submit a case for consideration, please email the relevant materials and presenting information to [email protected]. A 10-Year-Old Girl with Foot Pain After Falling from a Tree Figure 1. Figure 2. Case A 10-year-old girl presents with pain after falling from a tree, landing on her right foot. On examination, the pain emanates from the second through fifth metatarsals and proxi- mal phalanges. View the images taken and con- sider what the diagnosis and next steps would be. Resolution of the case is described on the next page. www.jucm.com JUCM The Journal of Urgent Care Medicine | February 2019 37 INSIGHTS IN IMAGES: CLINICAL CHALLENGE THE RESOLUTION Figure 1. Mediastinal air Figure 1. Differential Diagnosis Pearls for Urgent Care Management and Ⅲ Fracture of the distal fourth metatarsal Considerations for Transfer Ⅲ Plantar plate disruption Ⅲ Emergent transfer should be considered with associated neu- Ⅲ Sesamoiditis rologic deficit, compartment syndrome, open fracture, or vas- Ⅲ Turf toe cular compromise Ⅲ Referral to an orthopedist is warranted in the case of an in- Diagnosis tra-articular fracture, or with Lisfranc ligament injury or ten- Angulation of the distal fourth metatarsal metaphyseal cortex derness over the Lisfranc ligament and hairline lucency consistent with fracture. Acknowledgment: Images courtesy of Teleradiology Associates. Learnings/What to Look for Ⅲ Proximal metatarsal fractures are most often caused by crush- ing or direct blows Ⅲ In athletes, an axial load placed on a plantar-flexed foot should raise suspicion of a Lisfranc injury 38 JUCM The Journal of Urgent Care Medicine | February 2019 www.jucm.com INSIGHTS IN IMAGES CLINICAL CHALLENGE: CASE 2 A 55-Year-Old Man with 3 Hours of Epigastric Pain 55 years PR 249 QRSD 90 QT 471 QTc 425 AXES P 64 QRS -35 T 30 Figure 1. -

Genetic Mechanisms of Disease in Children: a New Look
Genetic Mechanisms of Disease in Children: A New Look Laurie Demmer, MD Tufts Medical Center and the Floating Hospital for Children Boston, MA Traditionally genetic disorders have been linked to the ‘one gene-one protein-one disease’ hypothesis. However recent advances in the field of molecular biology and biotechnology have afforded us the opportunity to greatly expand our knowledge of genetics, and we now know that the mechanisms of inherited disorders are often significantly more complex, and consequently, much more intriguing, than originally thought. Classical mendelian disorders with relatively simple genetic mechanisms do exist, but turn out to be far more rare than originally thought. All patients with sickle cell disease for example, carry the same A-to-T point mutation in the sixth codon of the beta globin gene. This results in a glutamate to valine substitution which changes the shape and the function of the globin molecule in a predictable way. Similarly, all patients with achondroplasia have a single base pair substitution at nucleotide #1138 of the FGFR3 gene. On the other hand, another common inherited disorder, cystic fibrosis, is known to result from changes in a specific transmembrane receptor (CFTR), but over 1000 different disease-causing mutations have been reported in this single gene. Since most commercial labs only test for between 23-100 different mutations, interpreting CFTR mutation testing is significantly complicated by the known risk of false negative results. Many examples of complex, or non-Mendelian, inheritance are now known to exist and include disorders of trinucleotide repeats, errors in imprinting, and gene dosage effects. -

Phenotypic and Genotypic Characterisation of Noonan-Like
1of5 ELECTRONIC LETTER J Med Genet: first published as 10.1136/jmg.2004.024091 on 2 February 2005. Downloaded from Phenotypic and genotypic characterisation of Noonan-like/ multiple giant cell lesion syndrome J S Lee, M Tartaglia, B D Gelb, K Fridrich, S Sachs, C A Stratakis, M Muenke, P G Robey, M T Collins, A Slavotinek ............................................................................................................................... J Med Genet 2005;42:e11 (http://www.jmedgenet.com/cgi/content/full/42/2/e11). doi: 10.1136/jmg.2004.024091 oonan-like/multiple giant cell lesion syndrome (NL/ MGCLS; OMIM 163955) is a rare condition1–3 with Key points Nphenotypic overlap with Noonan’s syndrome (OMIM 163950) and cherubism (OMIM 118400) (table 1). N Noonan-like/multiple giant cell lesion syndrome (NL/ Recently, missense mutations in the PTPN11 gene on MGCLS) has clinical similarities with Noonan’s syn- chromosome 12q24.1 have been identified as the cause of drome and cherubism. It is unclear whether it is a Noonan’s syndrome in 45% of familial and sporadic cases,45 distinct entity or a variant of Noonan’s syndrome or indicating genetic heterogeneity within the syndrome. In the cherubism. 5 study by Tartaglia et al, there was a family in which three N Three unrelated patients with NL/MGCLS were char- members had features of Noonan’s syndrome; two of these acterised, two of whom were found to have mutations had incidental mandibular giant cell lesions.3 All three in the PTPN11 gene, the mutation found in 45% of members were found to have a PTPN11 mutation known to patients with Noonan’s syndrome. -

Megalencephaly and Macrocephaly
277 Megalencephaly and Macrocephaly KellenD.Winden,MD,PhD1 Christopher J. Yuskaitis, MD, PhD1 Annapurna Poduri, MD, MPH2 1 Department of Neurology, Boston Children’s Hospital, Boston, Address for correspondence Annapurna Poduri, Epilepsy Genetics Massachusetts Program, Division of Epilepsy and Clinical Electrophysiology, 2 Epilepsy Genetics Program, Division of Epilepsy and Clinical Department of Neurology, Fegan 9, Boston Children’s Hospital, 300 Electrophysiology, Department of Neurology, Boston Children’s Longwood Avenue, Boston, MA 02115 Hospital, Boston, Massachusetts (e-mail: [email protected]). Semin Neurol 2015;35:277–287. Abstract Megalencephaly is a developmental disorder characterized by brain overgrowth secondary to increased size and/or numbers of neurons and glia. These disorders can be divided into metabolic and developmental categories based on their molecular etiologies. Metabolic megalencephalies are mostly caused by genetic defects in cellular metabolism, whereas developmental megalencephalies have recently been shown to be caused by alterations in signaling pathways that regulate neuronal replication, growth, and migration. These disorders often lead to epilepsy, developmental disabilities, and Keywords behavioral problems; specific disorders have associations with overgrowth or abnor- ► megalencephaly malities in other tissues. The molecular underpinnings of many of these disorders are ► hemimegalencephaly now understood, providing insight into how dysregulation of critical pathways leads to ► -

Cardiomyopathy Precision Panel Overview Indications
Cardiomyopathy Precision Panel Overview Cardiomyopathies are a group of conditions with a strong genetic background that structurally hinder the heart to pump out blood to the rest of the body due to weakness in the heart muscles. These diseases affect individuals of all ages and can lead to heart failure and sudden cardiac death. If there is a family history of cardiomyopathy it is strongly recommended to undergo genetic testing to be aware of the family risk, personal risk, and treatment options. Most types of cardiomyopathies are inherited in a dominant manner, which means that one altered copy of the gene is enough for the disease to present in an individual. The symptoms of cardiomyopathy are variable, and these diseases can present in different ways. There are 5 types of cardiomyopathies, the most common being hypertrophic cardiomyopathy: 1. Hypertrophic cardiomyopathy (HCM) 2. Dilated cardiomyopathy (DCM) 3. Restrictive cardiomyopathy (RCM) 4. Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) 5. Isolated Left Ventricular Non-Compaction Cardiomyopathy (LVNC). The Igenomix Cardiomyopathy Precision Panel serves as a diagnostic and tool ultimately leading to a better management and prognosis of the disease. It provides a comprehensive analysis of the genes involved in this disease using next-generation sequencing (NGS) to fully understand the spectrum of relevant genes. Indications The Igenomix Cardiomyopathy Precision Panel is indicated in those cases where there is a clinical suspicion of cardiomyopathy with or without the following manifestations: - Shortness of breath - Fatigue - Arrythmia (abnormal heart rhythm) - Family history of arrhythmia - Abnormal scans - Ventricular tachycardia - Ventricular fibrillation - Chest Pain - Dizziness - Sudden cardiac death in the family 1 Clinical Utility The clinical utility of this panel is: - The genetic and molecular diagnosis for an accurate clinical diagnosis of a patient with personal or family history of cardiomyopathy, channelopathy or sudden cardiac death. -

WES Gene Package Multiple Congenital Anomalie.Xlsx
Whole Exome Sequencing Gene package Multiple congenital anomalie, version 5, 1‐2‐2018 Technical information DNA was enriched using Agilent SureSelect Clinical Research Exome V2 capture and paired‐end sequenced on the Illumina platform (outsourced). The aim is to obtain 8.1 Giga base pairs per exome with a mapped fraction of 0.99. The average coverage of the exome is ~50x. Duplicate reads are excluded. Data are demultiplexed with bcl2fastq Conversion Software from Illumina. Reads are mapped to the genome using the BWA‐MEM algorithm (reference: http://bio‐bwa.sourceforge.net/). Variant detection is performed by the Genome Analysis Toolkit HaplotypeCaller (reference: http://www.broadinstitute.org/gatk/). The detected variants are filtered and annotated with Cartagenia software and classified with Alamut Visual. It is not excluded that pathogenic mutations are being missed using this technology. At this moment, there is not enough information about the sensitivity of this technique with respect to the detection of deletions and duplications of more than 5 nucleotides and of somatic mosaic mutations (all types of sequence changes). HGNC approved Phenotype description including OMIM phenotype ID(s) OMIM median depth % covered % covered % covered gene symbol gene ID >10x >20x >30x A4GALT [Blood group, P1Pk system, P(2) phenotype], 111400 607922 101 100 100 99 [Blood group, P1Pk system, p phenotype], 111400 NOR polyagglutination syndrome, 111400 AAAS Achalasia‐addisonianism‐alacrimia syndrome, 231550 605378 73 100 100 100 AAGAB Keratoderma, palmoplantar, -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Description of the Vanseq Subpanels Offered at Seattle Children’S Hospital
Description of the VANseq subpanels offered at Seattle Children’s Hospital Capillary Malformations (4 genes): EPHB4, GNA11, GNAQ, RASA1 Capillary malformation-arteriovenous malformation (CM-AVM) syndrome is associated with multiple small (1-2 cm diameter) capillary malformations and is due to loss of function mutations in EPHB4 or RASA1. Approximately 20% of individuals have AVMs, which can be life-threatening. Other features (telangiectasias, lymphedema, non- immune hydrops) have been associated. RASA1 and EPHB4 mutations are also associated with Parkes-Weber syndrome. Somatic activating mutations at codon 183 in the GNAQ gene cause isolated capillary malformations or Sturge Weber Syndrome. Somatic activating mutations at the same residue in GNA11 cause capillary malformation with overgrowth. LM/VM/AVM (17 genes) - Lymphatic Malformations, Venous Malformations, Arteriovenous Malformations: ACVRL1, ARAF, BRAF, ELMO2, ENG, EPHB4, GDF2, GLMN, HRAS, KRAS, MAP2K1, MAP3K3, NRAS, PIK3CA, PTEN, RASA1, TEK (TIE2) Most individuals with isolated lymphatic, venous, or arteriovenous malformations possess somatic, activating mutations in genes associated with cell growth and division. For many of these conditions, sequencing of affected, lesional tissue is required for mutation detection, and coordination with pathology is required. Although there are strong gene-phenotype correlations within this group, there is increasing recognition of phenotypic expansion and overlap. The most commonly mutated gene in this group of conditions is PIK3CA. • ~80% of isolated lymphatic malformations have pathogenic, tissue restricted variant in PIK3CA. • Most venous malformations have activating mutations in TEK (TIE2). TEK mutations can be isolated and somatic or multifocal, inherited in a dominant fashion. • Activating, somatic mutations in MAP2K1 are primarily associated with isolated extracranial AVMs. -

Fetal Hydrops in Combination with Gonadoblastoid Testicular Dysplasia May Represent a Lethal Type of Noonan Syndrome
etics & E en m G b ry n o a l o Reischer et al., Human Genet Embryol 2016, 6:3 m g u y H Human Genetics & Embryology DOI: 10.4172/2161-0436.1000137 ISSN: 2161-0436 Research Article Open Access Fetal Hydrops in Combination with Gonadoblastoid Testicular Dysplasia May Represent a Lethal Type of Noonan Syndrome Theresa Reischer1, Maximilian Schmid1, Sukirthini Balendran1, Manuel Nistal2, Julia Vodopiutz3, Elisabeth Krampl-Bettelheim4, Niko Popitsch5, Sandra Liebmann-Reindl6 and Berthold Streubel6,7* 1Department of Obstetrics and Feto-Maternal Medicine, Medical University of Vienna, Vienna, Austria 2Department of Pathology, Hospital La Paz, Universidad Autónoma de Madrid, Spain 3Department of Pediatrics, Medical University of Vienna, Vienna, Austria 4Fetomed - Center for Fetal Medicine, Doebling Private Hospital, Vienna, Austria 5Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK 6Core Facility Genomics, Medical University of Vienna, Vienna, Austria 7Department of Pathology, Medical University of Vienna, Austria Abstract Objectives: Fetal gonadoblastoid testicular dysplasia is a rare finding. The combination with typical features of Noonan syndrome has never been described so far. We performed genetic testing including whole exome sequencing in two cases with fetal hydrops, congenital heart disease and gonadoblastoid testicular dysplasia. Methods: Exome sequencing was performed in the index case, where high quality DNA was isolated from fetal blood. In the second case and in five further gonadoblastoma samples, conventional Sanger sequencing was performed on DNA isolated from formalin fixed, paraffin embedded tissue. Results: Whole exome sequencing of the index case revealed a pathogenic mutation in the RIT1 gene (c.270G>A (p.Met90Ile)), leading to the diagnosis of Noonan syndrome type 8. -

Costello Syndrome
orphananesthesia Anaesthesia recommendations for patients suffering from Costello syndrome Disease name: Costello syndrome ICD 10: Q87.8 Synonyms: Significant phenotypical overlap with CFC (cardiofaciocutaneous syndrome) and Noonan syndrome. Disease summary: Costello syndrome (CS) is a rare disorder (so-called RAS-opathy, see below), affecting up to 300 people worldwide. First described by Dr Jack Costello in 1977, the syndrome is characterised by failure to thrive (FTT), poor feeding, short stature, developmental delay, distinctive facial features, excessive loose skin, cardiac abnormalities, and an increased risk of tumour development. RAS is a family of genes coding for small GTPases and includes amongst others HRAS. The HRAS gene is a proto-oncogene, which forms part of the MAPK (mitogen activated protein kinase) signalling pathway. Up-regulation of this signalling pathway causes unopposed cell growth, causing tumour predisposition. The MAPK pathway is also the site of mutations causing both CFC and Noonan syndrome. CS can be caused by a number of mutations in the HRAS gene. Most mutations do occur de novo, but there is some evidence that a minority are inherited in an autosomal dominant manner. Patients with CS are born large for gestational age, and there is a strong association with polyhydramnios and preterm labour. Growth later slows due to feeding difficulties. Head circumference is affected to a lesser degree than height and weight, which gives rise to relative macrocephaly. Growth Hormone (GH) deficiency can cause neonatal hypoglycaemia, and contributes to growth retardation. The disease is characterised by distinctive facial features including downslanting palpebral fissures, epicanthic folds, ptosis, flattened nasal bridge (hypertelorism), low set ears, thick lips, macroglossia and short neck. -

Workshop Schedule
Workshop Schedule Friday, August 27th Registration: 4:00‐6:30 pm Dinner 6:30‐8:00 pm Reception 8:00‐10:00 Saturday, August 28th 7:00‐8:15 am Breakfast: 8:15am Welcome and Introductions 8:30‐10:15 am Session 1: Novel strategies to understand the causes/mechanisms of birth defects (Moderator: Angela Lin) 8:30 am Les Biesecker ‐ Using massively parallel sequencing technologies to provide improved molecular delineation of human malformation syndromes 9:15 am Bamshad ‐ Exome sequencing identifies a gene for Kabuki syndrome 9:30 am Boycott ‐ Next‐Generation sequencing strategies give insight into the mechanism of birth defects in a Canadian isolated population 9:45 am Bleyl ‐ Comparison of pooled allelic ratios (CoPAR) analysis: an efficient method for mapping genetic traits in extended pedigrees 10:00 am Allanson – Nablus mask‐like facial syndrome and blepharo‐naso‐facial syndrome are the same entity. Refinement of the critical region of chromosome 8q22.1 points to a potential candidate gene 10:15‐10:45 am BREAK 10:45‐12:00 pm Session 2: Novel strategies to understand the causes/mechanisms of birth defects (cont.) (Moderator: Mike Bamshad) 10:45 am Krantz ‐ Applying novel genomic tools towards understanding an old chromosomal diagnosis: Using genome‐wide expression and SNP genotyping to identify the true cause of Pallister‐Killian syndrome 11:00 am Paciorkowski ‐ Bioinformatic analysis of published and novel copy number variants suggests candidate genes and networks for infantile spasms 11:15 am Bernier ‐ Identification of a novel Fibulin -

Ashkenazi Jewish
Reproductive Genetic Carrier Screening in Canada All couples planning their families should have a three-generation family history taken, ideally in the preconception period. Attention should be paid to the red flags in Box 1 to assess risk to future offspring. A personal or family history of: congenital anomaly e.g. congenital heart defect, neural tube defect intellectual disability or developmental delay genetic syndrome e.g. neurofibromatosis, Noonan syndrome chromosomal disorder e.g. Down syndrome (trisomy 21), familial translocation muscular disorder e.g. X-linked Duchenne and Becker muscular dystrophies bleeding disorder e.g. X-linked hemophilia A or B stillbirth sudden unexplained death other major health concerns such as cardiomyopathy, neurological disease, epilepsy, hearing loss, autism, and psychiatric disorders consanguinity Box 1. Personal and family history red flags that should prompt a referral for genetic consultation, ideally when individuals are planning a family (preconception). A history of any of these red flags should prompt referral for genetic consultation. Individuals and their partners should be encouraged to make their best efforts to obtain confirmatory information such as medical records, genetic test results, even family photos. One’s ethnicity is an important piece of risk assessment as some populations are known to have a higher incidence of certain genetic conditions due to a founder effect. Founder effect confers reduced genetic diversity in a population descended from a small number of ancestors. Founder mutations refer to specific gene mutations observed at high frequency in a specific population due to the presence of that gene mutation in a single or small number of ancestors.