Mutations in the Pantothenate Kinase of Plasmodium Falciparum Confer Diverse Sensitivity Profiles to Antiplasmodial Pantothenate Analogues
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma Divulgatum
microorganisms Article Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma divulgatum Rafael Bargiela 1 , Karin Lanthaler 1,2, Colin M. Potter 1,2 , Manuel Ferrer 3 , Alexander F. Yakunin 1,2, Bela Paizs 1,2, Peter N. Golyshin 1,2 and Olga V. Golyshina 1,2,* 1 School of Natural Sciences, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK; [email protected] (R.B.); [email protected] (K.L.); [email protected] (C.M.P.); [email protected] (A.F.Y.); [email protected] (B.P.); [email protected] (P.N.G.) 2 Centre for Environmental Biotechnology, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK 3 Systems Biotechnology Group, Department of Applied Biocatalysis, CSIC—Institute of Catalysis, Marie Curie 2, 28049 Madrid, Spain; [email protected] * Correspondence: [email protected]; Tel.: +44-1248-388607; Fax: +44-1248-382569 Received: 27 April 2020; Accepted: 15 May 2020; Published: 19 May 2020 Abstract: The archaeon Cuniculiplasma divulgatum is ubiquitous in acidic environments with low-to-moderate temperatures. However, molecular mechanisms underlying its ability to thrive at lower temperatures remain unexplored. Using mass spectrometry (MS)-based proteomics, we analysed the effect of short-term (3 h) exposure to cold. The C. divulgatum genome encodes 2016 protein-coding genes, from which 819 proteins were identified in the cells grown under optimal conditions. In line with the peptidolytic lifestyle of C. divulgatum, its intracellular proteome revealed the abundance of proteases, ABC transporters and cytochrome C oxidase. From 747 quantifiable polypeptides, the levels of 582 proteins showed no change after the cold shock, whereas 104 proteins were upregulated suggesting that they might be contributing to cold adaptation. -

Table S1. List of Oligonucleotide Primers Used
Table S1. List of oligonucleotide primers used. Cla4 LF-5' GTAGGATCCGCTCTGTCAAGCCTCCGACC M629Arev CCTCCCTCCATGTACTCcgcGATGACCCAgAGCTCGTTG M629Afwd CAACGAGCTcTGGGTCATCgcgGAGTACATGGAGGGAGG LF-3' GTAGGCCATCTAGGCCGCAATCTCGTCAAGTAAAGTCG RF-5' GTAGGCCTGAGTGGCCCGAGATTGCAACGTGTAACC RF-3' GTAGGATCCCGTACGCTGCGATCGCTTGC Ukc1 LF-5' GCAATATTATGTCTACTTTGAGCG M398Arev CCGCCGGGCAAgAAtTCcgcGAGAAGGTACAGATACGc M398Afwd gCGTATCTGTACCTTCTCgcgGAaTTcTTGCCCGGCGG LF-3' GAGGCCATCTAGGCCATTTACGATGGCAGACAAAGG RF-5' GTGGCCTGAGTGGCCATTGGTTTGGGCGAATGGC RF-3' GCAATATTCGTACGTCAACAGCGCG Nrc2 LF-5' GCAATATTTCGAAAAGGGTCGTTCC M454Grev GCCACCCATGCAGTAcTCgccGCAGAGGTAGAGGTAATC M454Gfwd GATTACCTCTACCTCTGCggcGAgTACTGCATGGGTGGC LF-3' GAGGCCATCTAGGCCGACGAGTGAAGCTTTCGAGCG RF-5' GAGGCCTGAGTGGCCTAAGCATCTTGGCTTCTGC RF-3' GCAATATTCGGTCAACGCTTTTCAGATACC Ipl1 LF-5' GTCAATATTCTACTTTGTGAAGACGCTGC M629Arev GCTCCCCACGACCAGCgAATTCGATagcGAGGAAGACTCGGCCCTCATC M629Afwd GATGAGGGCCGAGTCTTCCTCgctATCGAATTcGCTGGTCGTGGGGAGC LF-3' TGAGGCCATCTAGGCCGGTGCCTTAGATTCCGTATAGC RF-5' CATGGCCTGAGTGGCCGATTCTTCTTCTGTCATCGAC RF-3' GACAATATTGCTGACCTTGTCTACTTGG Ire1 LF-5' GCAATATTAAAGCACAACTCAACGC D1014Arev CCGTAGCCAAGCACCTCGgCCGAtATcGTGAGCGAAG D1014Afwd CTTCGCTCACgATaTCGGcCGAGGTGCTTGGCTACGG LF-3' GAGGCCATCTAGGCCAACTGGGCAAAGGAGATGGA RF-5' GAGGCCTGAGTGGCCGTGCGCCTGTGTATCTCTTTG RF-3' GCAATATTGGCCATCTGAGGGCTGAC Kin28 LF-5' GACAATATTCATCTTTCACCCTTCCAAAG L94Arev TGATGAGTGCTTCTAGATTGGTGTCggcGAAcTCgAGCACCAGGTTG L94Afwd CAACCTGGTGCTcGAgTTCgccGACACCAATCTAGAAGCACTCATCA LF-3' TGAGGCCATCTAGGCCCACAGAGATCCGCTTTAATGC RF-5' CATGGCCTGAGTGGCCAGGGCTAGTACGACCTCG -

Pantothenate Kinase-Associated Neurodegeneration
Pantothenate kinase-associated neurodegeneration Description Pantothenate kinase-associated neurodegeneration (formerly called Hallervorden-Spatz syndrome) is a disorder of the nervous system. This condition is characterized by progressive difficulty with movement, typically beginning in childhood. Movement abnormalities include involuntary muscle spasms, rigidity, and trouble with walking that worsens over time. Many people with this condition also develop problems with speech ( dysarthria), and some develop vision loss. Additionally, affected individuals may experience a loss of intellectual function (dementia) and psychiatric symptoms such as behavioral problems, personality changes, and depression. Pantothenate kinase-associated neurodegeneration is characterized by an abnormal buildup of iron in certain areas of the brain. A particular change called the eye-of-the- tiger sign, which indicates an accumulation of iron, is typically seen on magnetic resonance imaging (MRI) scans of the brain in people with this disorder. Researchers have described classic and atypical forms of pantothenate kinase- associated neurodegeneration. The classic form usually appears in early childhood, causing severe problems with movement that worsen rapidly. Features of the atypical form appear later in childhood or adolescence and progress more slowly. Signs and symptoms vary, but the atypical form is more likely than the classic form to involve speech defects and psychiatric problems. A condition called HARP (hypoprebetalipoproteinemia, acanthocytosis, retinitis pigmentosa, and pallidal degeneration) syndrome, which was historically described as a separate syndrome, is now considered part of pantothenate kinase-associated neurodegeneration. Frequency The precise incidence of this condition is unknown. It is estimated to affect 1 to 3 per million people worldwide. Causes Mutations in the PANK2 gene cause pantothenate kinase-associated neurodegeneration. -

A Novel Nonsense Mutation in PANK2 Gene in Two Patients with Pantothenate Kinase-Associated Neurodegeneration
IJMCM Case Report Autumn 2016, Vol 5, No 4 A Novel Nonsense Mutation in PANK2 Gene in Two Patients with Pantothenate Kinase-Associated Neurodegeneration Soudeh Ghafouri-Fard 1, Vahid Reza Yassaee 2, Alireza Rezayi 3, Feyzollah Hashemi-Gorji 2, Nasrin Alipour 2, ∗ Mohammad Miryounesi 2 1. Department of Medical Genetics, Shahid Beheshti University of Medical sciences, Tehran, Iran. 2. Genomic Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran. 3. Pediatric Neurology Department, Loghman Hospital, Faculty of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran. Submmited 29 June 2016; Accepted 14 August 2016; Published 23 October 2016 Pantothenate kinase- associated neurodegeneration (PKAN) syndrome is a rare autosomal recessive disorder characterized by progressive extrapyramidal dysfunction and iron accumulation in the brain and axonal spheroids in the central nervous system. It has been shown that the disorder is caused by mutations in PANK2 gene which codes for a mitochondrial enzyme participating in coenzyme A biosynthesis. Here we report two cases of classic PKAN syndrome with early onset of neurodegenerative disorder. Mutational analysis has revealed that both are homozygous for a novel nonsense mutation in PANK2 gene (c.T936A (p.C312X)). The high prevalence of consanguineous marriages in Iran raises the likelihood of occurrence of autosomal recessive disorders such as PKAN and necessitates proper premarital genetic counseling. Further research is needed to provide the data on the prevalence of PKAN and identification of common PANK2 mutations in Iranian population. Key words : PANK2 , pantothenate kinase-associated neurodegeneration, mutation antothenate kinase-associated neurodegen- weighted which is due to iron deposition in the Peration (PKAN) syndrome is a rare autosomal periphery (hypointensity) and necrosis on its central recessive disorder characterized by progressive part (hyperintensity) (2). -

Biochemical and Proteomic Profiling of Maize Endosperm Texture and Protein Quality Kyla J
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Theses, Dissertations, and Student Research in Agronomy and Horticulture Department Agronomy and Horticulture 7-2015 Biochemical and Proteomic Profiling of Maize Endosperm Texture and Protein Quality Kyla J. Morton University of Nebraska-Lincoln Follow this and additional works at: http://digitalcommons.unl.edu/agronhortdiss Part of the Agricultural Science Commons, Agronomy and Crop Sciences Commons, Plant Biology Commons, and the Plant Breeding and Genetics Commons Morton, Kyla J., "Biochemical and Proteomic Profiling of Maize Endosperm Texture and Protein Quality" (2015). Theses, Dissertations, and Student Research in Agronomy and Horticulture. 88. http://digitalcommons.unl.edu/agronhortdiss/88 This Article is brought to you for free and open access by the Agronomy and Horticulture Department at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Theses, Dissertations, and Student Research in Agronomy and Horticulture by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. BIOCHEMICAL AND PROTEOMIC PROFILING OF MAIZE ENDOSPERM TEXTURE AND PROTEIN QUALITY by Kyla J. Morton A DISSERTATION Presented to the Faculty of The Graduate College at the University of Nebraska In Partial Fulfillment of Requirements For the Degree of Doctor of Philosophy Major: Agronomy and Horticulture (Plant Breeding and Genetics) Under the Supervision of Professor David R. Holding Lincoln, Nebraska July, 2015 BIOCHEMICAL AND PROTEOMIC ANALYSIS OF MAIZE ENDOSPERM KERNEL TEXTURE AND PROTEIN QUALITY Kyla J. Morton, Ph.D. University of Nebraska, 2015 Advisor: David R. Holding The research described herein, focuses on the biochemical and proteomic analysis of the maize endosperm and what influences kernel texture. -

O O2 Enzymes Available from Sigma Enzymes Available from Sigma
COO 2.7.1.15 Ribokinase OXIDOREDUCTASES CONH2 COO 2.7.1.16 Ribulokinase 1.1.1.1 Alcohol dehydrogenase BLOOD GROUP + O O + O O 1.1.1.3 Homoserine dehydrogenase HYALURONIC ACID DERMATAN ALGINATES O-ANTIGENS STARCH GLYCOGEN CH COO N COO 2.7.1.17 Xylulokinase P GLYCOPROTEINS SUBSTANCES 2 OH N + COO 1.1.1.8 Glycerol-3-phosphate dehydrogenase Ribose -O - P - O - P - O- Adenosine(P) Ribose - O - P - O - P - O -Adenosine NICOTINATE 2.7.1.19 Phosphoribulokinase GANGLIOSIDES PEPTIDO- CH OH CH OH N 1 + COO 1.1.1.9 D-Xylulose reductase 2 2 NH .2.1 2.7.1.24 Dephospho-CoA kinase O CHITIN CHONDROITIN PECTIN INULIN CELLULOSE O O NH O O O O Ribose- P 2.4 N N RP 1.1.1.10 l-Xylulose reductase MUCINS GLYCAN 6.3.5.1 2.7.7.18 2.7.1.25 Adenylylsulfate kinase CH2OH HO Indoleacetate Indoxyl + 1.1.1.14 l-Iditol dehydrogenase L O O O Desamino-NAD Nicotinate- Quinolinate- A 2.7.1.28 Triokinase O O 1.1.1.132 HO (Auxin) NAD(P) 6.3.1.5 2.4.2.19 1.1.1.19 Glucuronate reductase CHOH - 2.4.1.68 CH3 OH OH OH nucleotide 2.7.1.30 Glycerol kinase Y - COO nucleotide 2.7.1.31 Glycerate kinase 1.1.1.21 Aldehyde reductase AcNH CHOH COO 6.3.2.7-10 2.4.1.69 O 1.2.3.7 2.4.2.19 R OPPT OH OH + 1.1.1.22 UDPglucose dehydrogenase 2.4.99.7 HO O OPPU HO 2.7.1.32 Choline kinase S CH2OH 6.3.2.13 OH OPPU CH HO CH2CH(NH3)COO HO CH CH NH HO CH2CH2NHCOCH3 CH O CH CH NHCOCH COO 1.1.1.23 Histidinol dehydrogenase OPC 2.4.1.17 3 2.4.1.29 CH CHO 2 2 2 3 2 2 3 O 2.7.1.33 Pantothenate kinase CH3CH NHAC OH OH OH LACTOSE 2 COO 1.1.1.25 Shikimate dehydrogenase A HO HO OPPG CH OH 2.7.1.34 Pantetheine kinase UDP- TDP-Rhamnose 2 NH NH NH NH N M 2.7.1.36 Mevalonate kinase 1.1.1.27 Lactate dehydrogenase HO COO- GDP- 2.4.1.21 O NH NH 4.1.1.28 2.3.1.5 2.1.1.4 1.1.1.29 Glycerate dehydrogenase C UDP-N-Ac-Muramate Iduronate OH 2.4.1.1 2.4.1.11 HO 5-Hydroxy- 5-Hydroxytryptamine N-Acetyl-serotonin N-Acetyl-5-O-methyl-serotonin Quinolinate 2.7.1.39 Homoserine kinase Mannuronate CH3 etc. -

Biochemical Characterization of Pantoate Kinase, a Novel Enzyme Necessary for Coenzyme a Biosynthesis in the Archaea
Biochemical Characterization of Pantoate Kinase, a Novel Enzyme Necessary for Coenzyme A Biosynthesis in the Archaea Hiroya Tomita,a Yuusuke Yokooji,a Takuya Ishibashi,a Tadayuki Imanaka,b,c and Haruyuki Atomia,c Department of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University Katsura, Nishikyo-ku, Kyoto, Japana; Department of Biotechnology, College of Life Sciences, Ritsumeikan University Noji-Higashi, Kusatsu, Shiga, Japanb; and JST, CREST, Sanbancho, Chiyoda-ku, Tokyo, Japanc Although bacteria and eukaryotes share a pathway for coenzyme A (CoA) biosynthesis, we previously clarified that most archaea utilize a distinct pathway for the conversion of pantoate to 4=-phosphopantothenate. Whereas bacteria/eukaryotes use pantothe- nate synthetase and pantothenate kinase (PanK), the hyperthermophilic archaeon Thermococcus kodakarensis utilizes two novel enzymes: pantoate kinase (PoK) and phosphopantothenate synthetase (PPS). Here, we report a detailed biochemical examina- tion of PoK from T. kodakarensis. Kinetic analyses revealed that the PoK reaction displayed Michaelis-Menten kinetics toward ATP, whereas substrate inhibition was observed with pantoate. PoK activity was not affected by the addition of CoA/acetyl-CoA. Interestingly, PoK displayed broad nucleotide specificity and utilized ATP, GTP, UTP, and CTP with comparable kcat/Km values. Sequence alignment of 27 PoK homologs revealed seven conserved residues with reactive side chains, and variant proteins were constructed for each residue. Activity was not detected when mutations were introduced to Ser104, Glu134, and Asp143, suggest- ing that these residues play vital roles in PoK catalysis. Kinetic analysis of the other variant proteins, with mutations S28A, H131A, R155A, and T186A, indicated that all four residues are involved in pantoate recognition and that Arg155 and Thr186 play important roles in PoK catalysis. -

07 Nov 2016 FINAL 1 Consensus Clinical Management Guidelines for Pantothenate Kinase-Associated Neurodegeneration (PKAN) Penelo
Consensus Clinical Management Guidelines for Pantothenate Kinase-Associated Neurodegeneration (PKAN) Penelope Hogarth1,2, Manju A. Kurian3, Allison Gregory1, Barbara Csányi3, Tamara Zagustin4, Tomasz Kmiec5, Patricia Wood6, Angelika Klucken7, Natale Scalise8, Francesca Sofia8, Thomas Klopstock9,10,11, Giovanna Zorzi12, Nardo Nardocci12, Susan J. Hayflick1,2* 1Department of Molecular & Medical Genetics, Oregon Health & Science University, Portland USA 2Department of Neurology, Oregon Health & Science University, Portland USA 3Molecular Neurosciences, Developmental Neurosciences Programme, UCL Institute of Child Health, London UK 4Department of Physiatry, Children’s Healthcare of Atlanta, Georgia USA 5Department of Child Neurology, The Children’s Memorial Health Institute, Warsaw Poland 6NBIA Disorders Association, El Cajon CA USA 7Hoffunngsbaum e.V., Velbert Germany 8AISNAF – Associazione Italiana Sindromi Neurodegenerative Da Accumulo Di Ferro, Rossano Italy 9Department of Neurology, Friedrich-Baur-Institute, Ludwig-Maximilians- University, Munich, Germany 10German Center for Neurodegenerative Diseases (DZNE), Munich, Germany 11Munich Cluster for Systems Neurology (SyNergy), Munich, Germany 07 Nov 2016 FINAL 1 12Department of Pediatric Neuroscience, IRCCS Foundation Neurological Institute C. Besta, Milan Italy * Correspondence to: Susan J. Hayflick, MD, Molecular & Medical Genetics, mailcode L103, Oregon Health & Science University, Portland OR 97239 503.494.7703 07 Nov 2016 FINAL 2 Introduction Pantothenate kinase-associated neurodegeneration (PKAN, formerly Hallervorden– Spatz syndrome, OMIM #234200) is the most common neurodegeneration with brain iron accumulation (NBIA) disorder, with an estimated incidence of 1-3 per million1 and accounting for about half of NBIA cases.2 As is the case for most rare disorders, evidence-based guidance for the clinical management of PKAN is limited, often relying instead on anecdotal evidence, case reports and small series studies. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

SUPPORTING INFORMATION for Regulation of Gene Expression By
SUPPORTING INFORMATION for Regulation of gene expression by the BLM helicase correlates with the presence of G4 motifs Giang Huong Nguyen1,2, Weiliang Tang3, Ana I. Robles1, Richard P. Beyer4, Lucas T. Gray5, Judith A. Welsh1, Aaron J. Schetter1, Kensuke Kumamoto1,6, Xin Wei Wang1, Ian D. Hickson2,7, Nancy Maizels5, 3,8 1 Raymond J. Monnat, Jr. and Curtis C. Harris 1Laboratory of Human Carcinogenesis, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, U.S.A; 2Department of Medical Oncology, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, University of Oxford, Oxford, U.K.; 3Department of Pathology, University of Washington, Seattle, WA U.S.A.; 4 Center for Ecogenetics and Environmental Health, University of Washington, Seattle, WA U.S.A.; 5Department of Immunology and Department of Biochemistry, University of Washington, Seattle, WA U.S.A.; 6Department of Organ Regulatory Surgery, Fukushima Medical University, Fukushima, Japan; 7Cellular and Molecular Medicine, Nordea Center for Healthy Aging, University of Copenhagen, Denmark; 8Department of Genome Sciences, University of WA, Seattle, WA U.S.A. SI Index: Supporting Information for this manuscript includes the following 19 items. A more detailed Materials and Methods section is followed by 18 Tables and Figures in order of their appearance in the manuscript text: 1) SI Materials and Methods 2) Figure S1. Study design and experimental workflow. 3) Figure S2. Immunoblot verification of BLM depletion from human fibroblasts. 4) Figure S3. PCA of mRNA and miRNA expression in BLM-depleted human fibroblasts. 5) Figure S4. qPCR confirmation of mRNA array data. 6) Table S1. BS patient and control detail. -
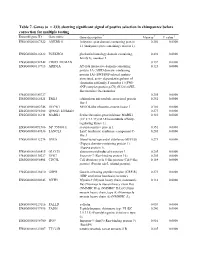
Table 7. Genes
Table 7. Genes (n = 233) showing significant signal of postive selection in chimpanzee before correction for multiple testing Ensembl gene ID Gene name Gene description * Mean ω † P value ‡ ENSG00000167522 ANKRD11 Ankyrin repeat domain-containing protein 0.256 0.0000 11 (Ankyrin repeat-containing cofactor 1). ENSG00000126822 PLEKHG3 pleckstrin homology domain containing, 0.492 0.0000 family G, member 3 ENSG00000152242 CR025_HUMAN 0.137 0.0000 ENSG00000117713 ARID1A AT-rich interactive domain-containing 0.123 0.0000 protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix- associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI- SNF complex protein p270) (B120) (SWI- like protein) (Osa homolog ENSG00000185727 0.205 0.0000 ENSG00000165521 EML5 echinoderm microtubule associated protein 0.252 0.0000 like 5 ENSG00000002746 HECW1 NEDD4-like ubiquitin-protein ligase 1 0.186 0.0000 ENSG00000198308 Q9NSI3_HUMAN 0.273 0.0000 ENSG00000116141 MARK1 Serine/threonine-protein kinase MARK1 0.410 0.0000 (EC 2.7.1.37) (MAP/microtubule affinity- regulating kinase 1). ENSG00000091986 NP_955806.1 steroid-sensitive protein 1 0.351 0.0000 ENSG00000147036 LANCL3 LanC lantibiotic synthetase component C- 0.205 0.0000 like 3 ENSG00000112276 BVES Blood vessel epicardial substance (hBVES) 0.273 0.0000 (Popeye domain-containing protein 1) (Popeye protein 1). ENSG00000106415 GLCCI1 glucocorticoid induced transcript 1 0.205 0.0000 ENSG00000130227 XPO7 Exportin-7 (Ran-binding protein 16). 0.205 0.0000 ENSG00000096401 CDC5L Cell division cycle 5-like protein (Cdc5-like 0.164 0.0000 protein) (Pombe cdc5- related protein). ENSG00000126010 GRPR Gastrin-releasing peptide receptor (GRP-R) 0.273 0.0000 (GRP-preferring bombesin receptor).