In Platelet Activation Prediction of Key Mediators Involved in Platelet Hyperreactivity Using a Novel Network Biology Approach
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -
Supplementary Table 1
Table S1. List of Genes Differentially Expressed in YB-1 siRNA-Transfected MCF-7 Cells Unigene Accession Symbol Description Mean fold change Hs.17466 NM_004585 RARRES3 retinoic acid receptor responder (tazarotene induced) 3 3.57 Hs.64746 NM_004669 CLIC3 chloride intracellular channel 3 3.33 Hs.516155 NM_001747 CAPG capping protein (actin filament), gelsolin-like 2.88 Hs.926 NM_002463 MX2 myxovirus (influenza virus) resistance 2 (mouse) 2.72 Hs.643494 NM_005410 SEPP1 selenoprotein P, plasma, 1 2.70 Hs.267038 BC017500 POF1B premature ovarian failure 1B 2.67 Hs.20961 U63917 GPR30 G protein- coupled receptor 30 2532.53 Hs.199877 BE645967 CPNE4 copine IV 2.49 Hs.632177 NM_017458 MVP major vault protein 2.48 Hs.528836 AA005023 NOD27 nucleotide-binding oligomerization domains 27 2.43 Hs.360940 AL589866 dJ222E13.1 kraken-like 2.40 Hs.515575 AI743780 PLAC2 placenta-specific 2 2.37 Hs.25674 AI827820 MBD2 methyl-CpG binding domain protein 2 2.36 Hs.12229 AA149594 TIEG2 TGFB inducible early growth response 2 2.33 Hs.482730 AA053711 EDIL3 EGF-like repeats and discoidin I-like domains 3 2.32 Hs. 370771 NM_000389 CDKN1A cyclin- depen dent kinase in hibitor 1A (p 21, Cip 1) 2282.28 Hs.469175 AI341537 JFC1 NADPH oxidase-related, C2 domain-containing protein 2.28 Hs.514821 AF043341 CCL5 chemokine (C-C motif) ligand 5 2.27 Hs.591292 NM_023915 GPR87 G protein-coupled receptor 87 2.26 Hs.500483 NM_001613 ACTA2 actin, alpha 2, smooth muscle, aorta 2.24 Hs.632824 NM_006729 DIAPH2 diaphanous homolog 2 (Drosophila) 2.22 Hs.369430 NM_000919 PAM peptidylglycine -

NICU Gene List Generator.Xlsx
Neonatal Crisis Sequencing Panel Gene List Genes: A2ML1 - B3GLCT A2ML1 ADAMTS9 ALG1 ARHGEF15 AAAS ADAMTSL2 ALG11 ARHGEF9 AARS1 ADAR ALG12 ARID1A AARS2 ADARB1 ALG13 ARID1B ABAT ADCY6 ALG14 ARID2 ABCA12 ADD3 ALG2 ARL13B ABCA3 ADGRG1 ALG3 ARL6 ABCA4 ADGRV1 ALG6 ARMC9 ABCB11 ADK ALG8 ARPC1B ABCB4 ADNP ALG9 ARSA ABCC6 ADPRS ALK ARSL ABCC8 ADSL ALMS1 ARX ABCC9 AEBP1 ALOX12B ASAH1 ABCD1 AFF3 ALOXE3 ASCC1 ABCD3 AFF4 ALPK3 ASH1L ABCD4 AFG3L2 ALPL ASL ABHD5 AGA ALS2 ASNS ACAD8 AGK ALX3 ASPA ACAD9 AGL ALX4 ASPM ACADM AGPS AMELX ASS1 ACADS AGRN AMER1 ASXL1 ACADSB AGT AMH ASXL3 ACADVL AGTPBP1 AMHR2 ATAD1 ACAN AGTR1 AMN ATL1 ACAT1 AGXT AMPD2 ATM ACE AHCY AMT ATP1A1 ACO2 AHDC1 ANK1 ATP1A2 ACOX1 AHI1 ANK2 ATP1A3 ACP5 AIFM1 ANKH ATP2A1 ACSF3 AIMP1 ANKLE2 ATP5F1A ACTA1 AIMP2 ANKRD11 ATP5F1D ACTA2 AIRE ANKRD26 ATP5F1E ACTB AKAP9 ANTXR2 ATP6V0A2 ACTC1 AKR1D1 AP1S2 ATP6V1B1 ACTG1 AKT2 AP2S1 ATP7A ACTG2 AKT3 AP3B1 ATP8A2 ACTL6B ALAS2 AP3B2 ATP8B1 ACTN1 ALB AP4B1 ATPAF2 ACTN2 ALDH18A1 AP4M1 ATR ACTN4 ALDH1A3 AP4S1 ATRX ACVR1 ALDH3A2 APC AUH ACVRL1 ALDH4A1 APTX AVPR2 ACY1 ALDH5A1 AR B3GALNT2 ADA ALDH6A1 ARFGEF2 B3GALT6 ADAMTS13 ALDH7A1 ARG1 B3GAT3 ADAMTS2 ALDOB ARHGAP31 B3GLCT Updated: 03/15/2021; v.3.6 1 Neonatal Crisis Sequencing Panel Gene List Genes: B4GALT1 - COL11A2 B4GALT1 C1QBP CD3G CHKB B4GALT7 C3 CD40LG CHMP1A B4GAT1 CA2 CD59 CHRNA1 B9D1 CA5A CD70 CHRNB1 B9D2 CACNA1A CD96 CHRND BAAT CACNA1C CDAN1 CHRNE BBIP1 CACNA1D CDC42 CHRNG BBS1 CACNA1E CDH1 CHST14 BBS10 CACNA1F CDH2 CHST3 BBS12 CACNA1G CDK10 CHUK BBS2 CACNA2D2 CDK13 CILK1 BBS4 CACNB2 CDK5RAP2 -
Thejournalofcellb Io Logy
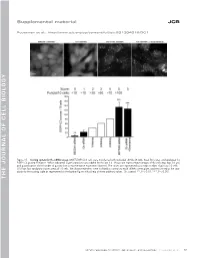
Supplemental material JCB Puustinen et al., http://www.jcb.org/cgi/content/full/jcb.201304012/DC1 Figure S1. Scoring system for the siRNA screen. MCF7-EGFP-LC3 cells were transfected with indicated siRNAs (8 nM), fixed 56 h later, and analyzed for EGFP-LC3 puncta formation. When indicated, 5 µM siramesine was added for the last 3 h. Shown are representative images of the cells (top; bar, 10 µm) and quantification of the number of puncta from a representative experiment (bottom). The values are represented as a mean number of puncta/10 cells ± SDs from four randomly chosen areas of 10 cells. The shown treatments were included as controls to each siRNA screen plate, and they served as the stan- dards for the scoring scale as represented on the bottom figure with a help of three arbitrary values. Ctr, control. **, P < 0.01; ***, P < 0.001. THE JOURNAL OF CELL BIOLOGY CIP2A regulates mTORC1, cell growth, and autophagy • Puustinen et al. S1 Figure S2. A cartoon highlighting the positions of the candidate autophagy-regulating genes in the insulin receptor signaling network. The cartoon was prepared by Ingenuity Pathway Analysis. software. CP, carbohydrate phosphatase; ERK, extracellular signal–regulated kinase; FSH, follicle-stimulating hor- mone; EG, epidermal growth factor. S2 JCB Figure S3. The effect of siRNAs targeting the autophagy-regulating PP2 subunits on target gene expression, cell density, and mTORC1 pathway. (A) Total RNA was isolated from MCF7 cells treated with indicated siRNAs (20 nM) for 54 h and analyzed for target gene expression by qPCR using primers de- signed to specifically amplify the indicated PP2A-related mRNAs andACTB mRNA (internal control). -

Phosphoproteomics of Retinoblastoma: a Pilot Study Identifies Aberrant Kinases
molecules Article Phosphoproteomics of Retinoblastoma: A Pilot Study Identifies Aberrant Kinases Lakshmi Dhevi Nagarajha Selvan 1,†, Ravikanth Danda 1,2,†, Anil K. Madugundu 3 ID , Vinuth N. Puttamallesh 3, Gajanan J. Sathe 3,4, Uma Maheswari Krishnan 2, Vikas Khetan 5, Pukhraj Rishi 5, Thottethodi Subrahmanya Keshava Prasad 3,6 ID , Akhilesh Pandey 3,7,8, Subramanian Krishnakumar 1, Harsha Gowda 3,* and Sailaja V. Elchuri 9,* 1 L&T Opthalmic Pathology, Vision Research Foundation, Sankara Nethralaya, Chennai, Tamil Nadu 600 006, India; [email protected] (L.D.N.S.); [email protected] (R.D.); [email protected] (S.K.) 2 Centre for Nanotechnology and Advanced Biomaterials, Shanmugha Arts, Science, Technology and Research Academy University, Tanjore, Tamil Nadu 613 401, India; [email protected] 3 Institute of Bioinformatics, International Technology Park, Bangalore, Karnataka 560 066, India; [email protected] (A.K.M.); [email protected] (V.N.P.); [email protected] (G.J.S.); [email protected] (T.S.K.P.); [email protected] (A.P.) 4 Manipal Academy of Higher Education (MAHE), Manipal, Karnataka 576 104, India 5 Shri Bhagwan Mahavir Vitreoretinal Services, Sankara Nethralaya, Chennai, Tamil Nadu 600 006, India; [email protected] (V.K.); [email protected] (P.R.) 6 Center for Systems Biology and Molecular Medicine, Yenepoya Research Centre, Yenepoya (Deemed to be University), Mangalore, Karnataka 575 108, India 7 McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA 8 Departments of Biological Chemistry, Pathology and Oncology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA 9 Department of Nanotechnology, Vision Research Foundation, Sankara Nethralaya, Chennai, Tamil Nadu 600 006, India * Correspondence: [email protected] (H.G.); [email protected] (S.V.E.); Tel.: +91-80-28416140 (H.G.); +91-44-28271616 (S.V.E.) † These authors contributed equally to this work. -

Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling
International Journal of Molecular Sciences Review Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling Maria Carmela Annunziata 1, Melania Parisi 1, Gabriella Esposito 2 , Gabriella Fabbrocini 1, Rosario Ammendola 2 and Fabio Cattaneo 2,* 1 Department of Clinical Medicine and Surgery, School of Medicine, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (M.C.A.); [email protected] (M.P.); [email protected] (G.F.) 2 Department of Molecular Medicine and Medical Biotechnology, School of Medicine,, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (G.E.); [email protected] (R.A.) * Correspondence: [email protected]; Fax: +39-081-7464-359 Received: 5 May 2020; Accepted: 25 May 2020; Published: 27 May 2020 Abstract: FPR1, FPR2, and FPR3 are members of Formyl Peptides Receptors (FPRs) family belonging to the GPCR superfamily. FPR2 is a low affinity receptor for formyl peptides and it is considered the most promiscuous member of this family. Intracellular signaling cascades triggered by FPRs include the activation of different protein kinases and phosphatase, as well as tyrosine kinase receptors transactivation. Protein kinases and phosphatases act coordinately and any impairment of their activation or regulation represents one of the most common causes of several human diseases. Several phospho-sites has been identified in protein kinases and phosphatases, whose role may be to expand the repertoire of molecular mechanisms of regulation or may be necessary for fine-tuning of switch properties. We previously performed a phospho-proteomic analysis in FPR2-stimulated cells that revealed, among other things, not yet identified phospho-sites on six protein kinases and one protein phosphatase. -

STRIPAK Complexes in Cell Signaling and Cancer
Oncogene (2016), 1–9 © 2016 Macmillan Publishers Limited All rights reserved 0950-9232/16 www.nature.com/onc REVIEW STRIPAK complexes in cell signaling and cancer Z Shi1,2, S Jiao1 and Z Zhou1,3 Striatin-interacting phosphatase and kinase (STRIPAK) complexes are striatin-centered multicomponent supramolecular structures containing both kinases and phosphatases. STRIPAK complexes are evolutionarily conserved and have critical roles in protein (de) phosphorylation. Recent studies indicate that STRIPAK complexes are emerging mediators and regulators of multiple vital signaling pathways including Hippo, MAPK (mitogen-activated protein kinase), nuclear receptor and cytoskeleton remodeling. Different types of STRIPAK complexes are extensively involved in a variety of fundamental biological processes ranging from cell growth, differentiation, proliferation and apoptosis to metabolism, immune regulation and tumorigenesis. Growing evidence correlates dysregulation of STRIPAK complexes with human diseases including cancer. In this review, we summarize the current understanding of the assembly and functions of STRIPAK complexes, with a special focus on cell signaling and cancer. Oncogene advance online publication, 15 February 2016; doi:10.1038/onc.2016.9 INTRODUCTION in the central nervous system and STRN4 is mostly abundant in Recent proteomic studies identified a group of novel multi- the brain and lung, whereas STRN3 is ubiquitously expressed in 5–9 component complexes named striatin (STRN)-interacting phos- almost all tissues. STRNs share a -

PHKB Gene Phosphorylase Kinase Regulatory Subunit Beta
PHKB gene phosphorylase kinase regulatory subunit beta Normal Function The PHKB gene provides instructions for making one piece, the beta subunit, of the phosphorylase b kinase enzyme. This enzyme is made up of 16 subunits, four each of the alpha, beta, gamma, and delta subunits. (Each subunit is produced from a different gene.) The beta subunit helps regulate the activity of phosphorylase b kinase. This enzyme is found in various tissues, although it is most abundant in the liver and muscles. One version of the enzyme is found in liver cells and another in muscle cells. The beta subunit produced from the PHKB gene is part of the enzyme found both in the liver and in muscle. Phosphorylase b kinase plays an important role in providing energy for cells. The main source of cellular energy is a simple sugar called glucose. Glucose is stored in muscle and liver cells in a form called glycogen. Glycogen can be broken down rapidly when glucose is needed, for instance during exercise. Phosphorylase b kinase turns on ( activates) another enzyme called glycogen phosphorylase b by converting it to the more active form, glycogen phosphorylase a. When active, this enzyme breaks down glycogen. Health Conditions Related to Genetic Changes Glycogen storage disease type IX At least 21 mutations in the PHKB gene are known to cause a form of glycogen storage disease type IX (GSD IX) called GSD IXb. This form of the disorder affects the liver and the muscles. The liver problems caused by this disorder include an enlarged liver ( hepatomegaly), slow growth, and periods of low blood sugar (hypoglycemia). -

Clinical, Biochemical, and Genetic Characterization of Glycogen Storage Type IX in a Child with Asymptomatic Hepatomegaly
pISSN: 2234-8646 eISSN: 2234-8840 http://dx.doi.org/10.5223/pghn.2015.18.2.138 Pediatr Gastroenterol Hepatol Nutr 2015 June 18(2):138-143 Case Report PGHN Clinical, Biochemical, and Genetic Characterization of Glycogen Storage Type IX in a Child with Asymptomatic Hepatomegaly Jung Ah Kim, Ja Hye Kim, Beom Hee Lee, Gu-Hwan Kim*, Yoon S. Shin†, Han-Wook Yoo, and Kyung Mo Kim Department of Pediatrics, *Medical Genetics Center, Asan Medical Center Children’s Hospital, University of Ulsan College of Medicine, Seoul, Korea, †University Children's Hospital and Molecular Genetics and Metabolism Laboratory, Munich, Germany Glycogen storage disease type IX (GSD IX) is caused by a defect in phosphorylase b kinase (PhK) that results from mutations in the PHKA2, PHKB, and PHKG2 genes. Patients usually manifest recurrent ketotic hypoglycemia with growth delay, but some may present simple hepatomegaly. Although GSD IX is one of the most common causes of GSDs, its biochemical and genetic diagnosis has been problematic due to its rarity, phenotypic overlap with other types of GSDs, and genetic heterogeneities. In our report, a 22-month-old boy with GSD IX is described. No other manifestations were evident except for hepatomegaly. His growth and development also have been proceeding normally. Diagnosed was made by histologic examination, an enzyme assay, and genetic testing with known c.3210_3212del (p.Arg1070del) mutation in PHKA2 gene. Key Words: Glycogen storage disease, Glycogen storage disease type IX, Phosphorylase b kinase 2, Phosphorylase kinase, Hepatomegaly INTRODUCTION lism [1]. GSD type IX (GSD IX) results from a defi- ciency of phosphorylase b kinase (PhK), which plays Glycogen storage disease (GSD) is a group of dis- an essential role in regulating the breakdown of gly- eases caused by inborn errors of glycogen metabo- cogen to glucose. -

Download 20190410); Fragmentation for 20 S
ARTICLE https://doi.org/10.1038/s41467-020-17387-y OPEN Multi-layered proteomic analyses decode compositional and functional effects of cancer mutations on kinase complexes ✉ Martin Mehnert 1 , Rodolfo Ciuffa1, Fabian Frommelt 1, Federico Uliana1, Audrey van Drogen1, ✉ ✉ Kilian Ruminski1,3, Matthias Gstaiger1 & Ruedi Aebersold 1,2 fi 1234567890():,; Rapidly increasing availability of genomic data and ensuing identi cation of disease asso- ciated mutations allows for an unbiased insight into genetic drivers of disease development. However, determination of molecular mechanisms by which individual genomic changes affect biochemical processes remains a major challenge. Here, we develop a multilayered proteomic workflow to explore how genetic lesions modulate the proteome and are trans- lated into molecular phenotypes. Using this workflow we determine how expression of a panel of disease-associated mutations in the Dyrk2 protein kinase alter the composition, topology and activity of this kinase complex as well as the phosphoproteomic state of the cell. The data show that altered protein-protein interactions caused by the mutations are asso- ciated with topological changes and affected phosphorylation of known cancer driver pro- teins, thus linking Dyrk2 mutations with cancer-related biochemical processes. Overall, we discover multiple mutation-specific functionally relevant changes, thus highlighting the extensive plasticity of molecular responses to genetic lesions. 1 Department of Biology, Institute of Molecular Systems Biology, ETH Zurich, -

The Mitotic Checkpoint Is a Targetable Vulnerability of Carboplatin-Resistant
www.nature.com/scientificreports OPEN The mitotic checkpoint is a targetable vulnerability of carboplatin‑resistant triple negative breast cancers Stijn Moens1,2, Peihua Zhao1,2, Maria Francesca Baietti1,2, Oliviero Marinelli2,3, Delphi Van Haver4,5,6, Francis Impens4,5,6, Giuseppe Floris7,8, Elisabetta Marangoni9, Patrick Neven2,10, Daniela Annibali2,11,13, Anna A. Sablina1,2,13 & Frédéric Amant2,10,12,13* Triple‑negative breast cancer (TNBC) is the most aggressive breast cancer subtype, lacking efective therapy. Many TNBCs show remarkable response to carboplatin‑based chemotherapy, but often develop resistance over time. With increasing use of carboplatin in the clinic, there is a pressing need to identify vulnerabilities of carboplatin‑resistant tumors. In this study, we generated carboplatin‑resistant TNBC MDA‑MB‑468 cell line and patient derived TNBC xenograft models. Mass spectrometry‑based proteome profling demonstrated that carboplatin resistance in TNBC is linked to drastic metabolism rewiring and upregulation of anti‑oxidative response that supports cell replication by maintaining low levels of DNA damage in the presence of carboplatin. Carboplatin‑ resistant cells also exhibited dysregulation of the mitotic checkpoint. A kinome shRNA screen revealed that carboplatin‑resistant cells are vulnerable to the depletion of the mitotic checkpoint regulators, whereas the checkpoint kinases CHEK1 and WEE1 are indispensable for the survival of carboplatin‑ resistant cells in the presence of carboplatin. We confrmed that pharmacological inhibition of CHEK1 by prexasertib in the presence of carboplatin is well tolerated by mice and suppresses the growth of carboplatin‑resistant TNBC xenografts. Thus, abrogation of the mitotic checkpoint by CHEK1 inhibition re‑sensitizes carboplatin‑resistant TNBCs to carboplatin and represents a potential strategy for the treatment of carboplatin‑resistant TNBCs. -

Human Kinases Info Page
Human Kinase Open Reading Frame Collecon Description: The Center for Cancer Systems Biology (Dana Farber Cancer Institute)- Broad Institute of Harvard and MIT Human Kinase ORF collection from Addgene consists of 559 distinct human kinases and kinase-related protein ORFs in pDONR-223 Gateway® Entry vectors. All clones are clonal isolates and have been end-read sequenced to confirm identity. Kinase ORFs were assembled from a number of sources; 56% were isolated as single cloned isolates from the ORFeome 5.1 collection (horfdb.dfci.harvard.edu); 31% were cloned from normal human tissue RNA (Ambion) by reverse transcription and subsequent PCR amplification adding Gateway® sequences; 11% were cloned into Entry vectors from templates provided by the Harvard Institute of Proteomics (HIP); 2% additional kinases were cloned into Entry vectors from templates obtained from collaborating laboratories. All ORFs are open (stop codons removed) except for 5 (MST1R, PTK7, JAK3, AXL, TIE1) which are closed (have stop codons). Detailed information can be found at: www.addgene.org/human_kinases Handling and Storage: Store glycerol stocks at -80oC and minimize freeze-thaw cycles. To access a plasmid, keep the plate on dry ice to prevent thawing. Using a sterile pipette tip, puncture the seal above an individual well and spread a portion of the glycerol stock onto an agar plate. To patch the hole, use sterile tape or a portion of a fresh aluminum seal. Note: These plasmid constructs are being distributed to non-profit institutions for the purpose of basic