Analysis of the Role of the Integrin Signaling Pathway in Hepatocytes During Rat Liver Regeneration
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PI3K Catalytic Isoform Alteration Promotes the LIMK1-Related
ANTICANCER RESEARCH 37 : 1805-1818 (2017) doi:10.21873/anticanres.11515 PI3K Catalytic Isoform Alteration Promotes the LIMK1-related Metastasis Through the PAK1 or ROCK1/2 Activation in Cigarette Smoke-exposed Ovarian Cancer Cells GA BIN PARK 1 and DAEJIN KIM 2 1Department of Biochemistry, Kosin University College of Medicine, Busan, Republic of Korea; 2Department of Anatomy, Inje University College of Medicine, Busan, Republic of Korea Abstract. Aim: To investigate the molecular mechanisms Several studies have shown a strong correlation between by which long-term exposure to cigarette smoke extract cigarette smoke (CS) and cancer metastasis through the (CSE) contributes to ovarian cancer metastasis. Materials induction of numerous factors involved in migration activity and Methods: Western blot analysis for diverse p110 (1-3). The exposure to CS induces the epithelial- isoforms of phosphoinositide 3-kinase (PI3K)-related mesenchymal transition (EMT) process and up-regulates the signaling pathway and epithelial-mesenchymal transition expression of EMT markers, including N-cadherin and (EMT) markers was performed to analyze the underlying vimentin (4, 5). Cigarette smoke extract (CSE) treatment mechanisms. Migratory activity of CSE-exposed ovarian significantly induces interleukin-8 (IL-8) and transforming cancer cells was determined by transendothelial migration growth factor-beta 1 (TGF- β1 ) production and profoundly and invasion assay. Results: After exposure to CSE for four suppresses the proliferation and growth of erythroid and weeks, CaOV3 (primary) and SKOV3 (metastatic) ovarian granulocyte-macrophage progenitors (6). Stimulation with cancer cells showed enhanced mesenchymal characteristics CSE in human lung fibroblast cells induces the expression and produced EMT-related cytokines [intwerleukin-8 (IL-8), of phosphorylated Smad3, a main downstream target of the vascular endothelial growth factor (VEGF) and TGF- β1 receptor, which results in the secretion of vascular transforming growth factor-beta 1 (TGF- β1 )]. -

Circular RNA Hsa Circ 0005114‑Mir‑142‑3P/Mir‑590‑5P‑ Adenomatous
ONCOLOGY LETTERS 21: 58, 2021 Circular RNA hsa_circ_0005114‑miR‑142‑3p/miR‑590‑5p‑ adenomatous polyposis coli protein axis as a potential target for treatment of glioma BO WEI1*, LE WANG2* and JINGWEI ZHAO1 1Department of Neurosurgery, China‑Japan Union Hospital of Jilin University, Changchun, Jilin 130033; 2Department of Ophthalmology, The First Hospital of Jilin University, Jilin University, Changchun, Jilin 130021, P.R. China Received September 12, 2019; Accepted October 22, 2020 DOI: 10.3892/ol.2020.12320 Abstract. Glioma is the most common type of brain tumor APC expression with a good overall survival rate. UALCAN and is associated with a high mortality rate. Despite recent analysis using TCGA data of glioblastoma multiforme and the advances in treatment options, the overall prognosis in patients GSE25632 and GSE103229 microarray datasets showed that with glioma remains poor. Studies have suggested that circular hsa‑miR‑142‑3p/hsa‑miR‑590‑5p was upregulated and APC (circ)RNAs serve important roles in the development and was downregulated. Thus, hsa‑miR‑142‑3p/hsa‑miR‑590‑5p‑ progression of glioma and may have potential as therapeutic APC‑related circ/ceRNA axes may be important in glioma, targets. However, the expression profiles of circRNAs and their and hsa_circ_0005114 interacted with both of these miRNAs. functions in glioma have rarely been studied. The present study Functional analysis showed that hsa_circ_0005114 was aimed to screen differentially expressed circRNAs (DECs) involved in insulin secretion, while APC was associated with between glioma and normal brain tissues using sequencing the Wnt signaling pathway. In conclusion, hsa_circ_0005114‑ data collected from the Gene Expression Omnibus database miR‑142‑3p/miR‑590‑5p‑APC ceRNA axes may be potential (GSE86202 and GSE92322 datasets) and explain their mecha‑ targets for the treatment of glioma. -

Oncogenic Activation of Pak1-Dependent Pathway of Macropinocytosis Determines BCG Entry Into Bladder Cancer Cells
Cancer Molecular and Cellular Pathobiology Research Oncogenic Activation of Pak1-Dependent Pathway of Macropinocytosis Determines BCG Entry into Bladder Cancer Cells Gil Redelman-Sidi1,2, Gopa Iyer3,4, David B. Solit3,4, and Michael S. Glickman1,2 Abstract Bacille Calmette-Guerin (BCG) is an attenuated strain of Mycobacterium bovis that is used widely as a vaccine for tuberculosis and is used as an effective treatment for superficial bladder carcinoma. Despite being the most successful cancer biotherapy, its mechanism of action and response determinants remain obscure. Here, we establish a model system to analyze BCG interaction with bladder cancer cells, using it to show that these cells vary dramatically in their susceptibility to BCG infection. Unexpectedly, the uptake of BCG by bladder cancer cells occurs by macropinocytosis rather than phagocytosis. BCG entry into bladder cancer cells relied upon Rac1, Cdc42, and their effector kinase Pak1. The difference in susceptibility between BCG- permissive and -resistant bladder cancer cells was due to oncogenic activation of signaling pathways that activate macropinocytosis, with phosphoinositide 3-kinase inhibitor activation stimulating BCG uptake independently of Akt. Similarly, activated Ras strongly activated Pak1-dependent uptake of BCG. These results reveal that oncogenic activation of macropinocytosis determines BCG uptake by bladder cancer cells, implying that tumor responsiveness to BCG may be governed by the specific mutations present in the treated cancer cell. Cancer Res; 73(3); 1156–67. Ó2013 AACR. Introduction Despite more than 30 years of clinical experience with Bladder cancer is among the most common tumors diag- intravesical BCG for bladder cancer, its mechanism of antitu- nosed in the United States, with an estimated annual incidence mor effect remains unknown and no markers exist to predict of 70,530 new cases and 14,680 deaths in 2010 (1). -

Role of Rac1-Pak Pathway in Aggressive B-Cell Lymphoma
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-4-2019 Role of Rac1-Pak pathway in aggressive b-cell lymphoma Tian Tian University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Hemic and Lymphatic Diseases Commons, and the Neoplasms Commons Recommended Citation Tian, Tian, "Role of Rac1-Pak pathway in aggressive b-cell lymphoma" (2019). Theses & Dissertations. 344. https://digitalcommons.unmc.edu/etd/344 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. i Role of Rac1-PAK Pathway in Aggressive B-cell Lymphomas By Tian Tian A DISSERTATION Presented to the Faculty of The University of Nebraska Graduate College in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Pathology & Microbiology Graduate program Under the Supervision of Professor Kai Fu University of Nebraska Medical Center, Omaha, Nebraska Dec, 2018 Supervisory Committee: Ying Yan, Ph.D. John S Davis, Ph.D. Timothy C Greiner, M.D. Javeed Iqbal, Ph.D. ii Role of Rac1-PAK pathway in Aggressive B-cell Lymphomas Tian Tian University of Nebraska Medical Center, 2018 Advisor: Kai Fu, M.D. Ph.D. Aggressive B-cell lymphomas are diverse group of neoplasms that arise at different stages of B-cell development and by various mechanisms of neoplastic transformation. Aggressive B-cell lymphomas include many types, subtypes and variants of diffuse large B-cell lymphoma (DLBCL), Burkitt lymphoma (BL), mantle cell lymphoma (MCL) and B lymphoblastic lymphoma. -

Whole Exome Sequencing of Thymic Neuroendocrine Tumor with Ectopic
176:2 Y Li, Y Peng and others Sequencing thymic 176:2 187–194 Clinical Study neuroendocrine tumor Whole exome sequencing of thymic neuroendocrine tumor with ectopic ACTH syndrome Yanli Li1,*, Ying Peng1,*, Xiuli Jiang1, Yulong Cheng3, Weiwei Zhou1, Tingwei Su1, Jing Xie2, Xu Zhong1, Dalong Song1, Luming Wu1, Liwen Fan1, Min Li1, Jie Hong1, Weiqing Wang1, Guang Ning1,3 and Yanan Cao1 1Shanghai Clinical Center for Endocrine and Metabolic Diseases, Shanghai Key Laboratory for Endocrine Tumors and 2Department of Pathology, Rui-Jin Hospital, Shanghai Jiao-Tong University School of Medicine, Shanghai, China, and 3Laboratory of Endocrinology and Metabolism, Institute of Health Correspondence Sciences, Shanghai Institutes for Biological Sciences (SIBS), Chinese Academy of Sciences (CAS) & should be addressed Shanghai Jiao-Tong University School of Medicine (SJTUSM), Shanghai, China to Y Cao *(Y Li and Y Peng contributed equally to this work) Email [email protected] Abstract Objective: Thymic neuroendocrine tumor is the second-most prevalent cause of ectopic adrenocorticotropic hormone (ACTH) syndrome (EAS), which is a rare disease characterized by ectopic ACTH oversecretion from nonpituitary tumors. However, the genetic abnormalities of thymic neuroendocrine tumors with EAS remain largely unknown. We aim to elucidate the genetic abnormalities and identify the somatic mutations of potential tumor-related genes of thymic neuroendocrine tumors with EAS by whole exome sequencing. Design and methods: Nine patients with thymic neuroendocrine tumors with EAS who were diagnosed at Shanghai Clinical Center for Endocrine and Metabolic Diseases in Ruijin Hospital between 2002 and 2014 were enrolled. We performed whole exome sequencing on the DNA obtained from thymic neuroendocrine tumors and matched peripheral blood using the Hiseq2000 platform. -

Supplementary Table S2
1-high in cerebrotropic Gene P-value patients Definition BCHE 2.00E-04 1 Butyrylcholinesterase PLCB2 2.00E-04 -1 Phospholipase C, beta 2 SF3B1 2.00E-04 -1 Splicing factor 3b, subunit 1 BCHE 0.00022 1 Butyrylcholinesterase ZNF721 0.00028 -1 Zinc finger protein 721 GNAI1 0.00044 1 Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 GNAI1 0.00049 1 Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 PDE1B 0.00069 -1 Phosphodiesterase 1B, calmodulin-dependent MCOLN2 0.00085 -1 Mucolipin 2 PGCP 0.00116 1 Plasma glutamate carboxypeptidase TMX4 0.00116 1 Thioredoxin-related transmembrane protein 4 C10orf11 0.00142 1 Chromosome 10 open reading frame 11 TRIM14 0.00156 -1 Tripartite motif-containing 14 APOBEC3D 0.00173 -1 Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D ANXA6 0.00185 -1 Annexin A6 NOS3 0.00209 -1 Nitric oxide synthase 3 SELI 0.00209 -1 Selenoprotein I NYNRIN 0.0023 -1 NYN domain and retroviral integrase containing ANKFY1 0.00253 -1 Ankyrin repeat and FYVE domain containing 1 APOBEC3F 0.00278 -1 Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F EBI2 0.00278 -1 Epstein-Barr virus induced gene 2 ETHE1 0.00278 1 Ethylmalonic encephalopathy 1 PDE7A 0.00278 -1 Phosphodiesterase 7A HLA-DOA 0.00305 -1 Major histocompatibility complex, class II, DO alpha SOX13 0.00305 1 SRY (sex determining region Y)-box 13 ABHD2 3.34E-03 1 Abhydrolase domain containing 2 MOCS2 0.00334 1 Molybdenum cofactor synthesis 2 TTLL6 0.00365 -1 Tubulin tyrosine ligase-like family, member 6 SHANK3 0.00394 -1 SH3 and multiple ankyrin repeat domains 3 ADCY4 0.004 -1 Adenylate cyclase 4 CD3D 0.004 -1 CD3d molecule, delta (CD3-TCR complex) (CD3D), transcript variant 1, mRNA. -
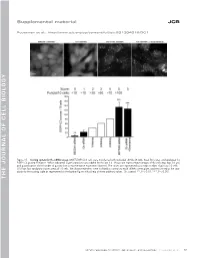
Thejournalofcellb Io Logy
Supplemental material JCB Puustinen et al., http://www.jcb.org/cgi/content/full/jcb.201304012/DC1 Figure S1. Scoring system for the siRNA screen. MCF7-EGFP-LC3 cells were transfected with indicated siRNAs (8 nM), fixed 56 h later, and analyzed for EGFP-LC3 puncta formation. When indicated, 5 µM siramesine was added for the last 3 h. Shown are representative images of the cells (top; bar, 10 µm) and quantification of the number of puncta from a representative experiment (bottom). The values are represented as a mean number of puncta/10 cells ± SDs from four randomly chosen areas of 10 cells. The shown treatments were included as controls to each siRNA screen plate, and they served as the stan- dards for the scoring scale as represented on the bottom figure with a help of three arbitrary values. Ctr, control. **, P < 0.01; ***, P < 0.001. THE JOURNAL OF CELL BIOLOGY CIP2A regulates mTORC1, cell growth, and autophagy • Puustinen et al. S1 Figure S2. A cartoon highlighting the positions of the candidate autophagy-regulating genes in the insulin receptor signaling network. The cartoon was prepared by Ingenuity Pathway Analysis. software. CP, carbohydrate phosphatase; ERK, extracellular signal–regulated kinase; FSH, follicle-stimulating hor- mone; EG, epidermal growth factor. S2 JCB Figure S3. The effect of siRNAs targeting the autophagy-regulating PP2 subunits on target gene expression, cell density, and mTORC1 pathway. (A) Total RNA was isolated from MCF7 cells treated with indicated siRNAs (20 nM) for 54 h and analyzed for target gene expression by qPCR using primers de- signed to specifically amplify the indicated PP2A-related mRNAs andACTB mRNA (internal control). -

Dysregulation of Rho Gtpases in the Apix/Arhgef6 Mouse Model of X
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2011 Dysregulation of Rho GTPases in the Pix/Arhgef6 mouse model of X-linked intellectual disability is paralleled by impaired structural and synaptic plasticity and cognitive deficits Ramakers, G J A ; Wolfer, D ; Rosenberger, G ; Kuchenbecker, K ; Kreienkamp, H J ; Prange-Kiel, J ; Rune, G ; Richter, K ; Langnaese, K ; Masneuf, S ; Bösl, M R ; Fischer, K D ; Krugers, H J ; Lipp, H P ; van Galen, E ; Kutsche, K Abstract: Mutations in the ARHGEF6 gene, encoding the guanine nucleotide exchange factor PIX/Cool- 2 for the Rho GTPases Rac1 and Cdc42, cause X-linked intellectual disability (ID) in humans. We show here that Pix/Arhgef6 is primarily expressed in neuropil regions of the hippocampus. To study the role of Pix/Arhgef6 in neuronal development and plasticity and gain insight into the pathogenic mechanisms underlying ID, we generated Pix/Arhgef6-deficient mice. Gross brain structure in these mice appeared to be normal; however, analysis of Golgi-Cox-stained pyramidal neurons revealed an increase in both dendritic length and spine density in the hippocampus, accompanied by an overall loss in spine synapses. Early-phase long-term potentiation was reduced and long-term depression was increased in the CA1 hippocampal area of Pix/Arhgef6-deficient animals. Knockout animals exhibited impaired spatial and complex learning and less behavioral control in mildly stressful situations, suggesting that this model mimics the human ID phenotype. The structural and electrophysiological alterations in the hippocampus were accompanied by a significant reduction in active Rac1 and Cdc42, but not RhoA. -
HCC and Cancer Mutated Genes Summarized in the Literature Gene Symbol Gene Name References*
HCC and cancer mutated genes summarized in the literature Gene symbol Gene name References* A2M Alpha-2-macroglobulin (4) ABL1 c-abl oncogene 1, receptor tyrosine kinase (4,5,22) ACBD7 Acyl-Coenzyme A binding domain containing 7 (23) ACTL6A Actin-like 6A (4,5) ACTL6B Actin-like 6B (4) ACVR1B Activin A receptor, type IB (21,22) ACVR2A Activin A receptor, type IIA (4,21) ADAM10 ADAM metallopeptidase domain 10 (5) ADAMTS9 ADAM metallopeptidase with thrombospondin type 1 motif, 9 (4) ADCY2 Adenylate cyclase 2 (brain) (26) AJUBA Ajuba LIM protein (21) AKAP9 A kinase (PRKA) anchor protein (yotiao) 9 (4) Akt AKT serine/threonine kinase (28) AKT1 v-akt murine thymoma viral oncogene homolog 1 (5,21,22) AKT2 v-akt murine thymoma viral oncogene homolog 2 (4) ALB Albumin (4) ALK Anaplastic lymphoma receptor tyrosine kinase (22) AMPH Amphiphysin (24) ANK3 Ankyrin 3, node of Ranvier (ankyrin G) (4) ANKRD12 Ankyrin repeat domain 12 (4) ANO1 Anoctamin 1, calcium activated chloride channel (4) APC Adenomatous polyposis coli (4,5,21,22,25,28) APOB Apolipoprotein B [including Ag(x) antigen] (4) AR Androgen receptor (5,21-23) ARAP1 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 (4) ARHGAP35 Rho GTPase activating protein 35 (21) ARID1A AT rich interactive domain 1A (SWI-like) (4,5,21,22,24,25,27,28) ARID1B AT rich interactive domain 1B (SWI1-like) (4,5,22) ARID2 AT rich interactive domain 2 (ARID, RFX-like) (4,5,22,24,25,27,28) ARID4A AT rich interactive domain 4A (RBP1-like) (28) ARID5B AT rich interactive domain 5B (MRF1-like) (21) ASPM Asp (abnormal -

The Mental Retardation Protein PAK3 Contributes to Synapse Formation and Plasticity in Hippocampus
10816 • The Journal of Neuroscience, December 1, 2004 • 24(48):10816–10825 Development/Plasticity/Repair The Mental Retardation Protein PAK3 Contributes to Synapse Formation and Plasticity in Hippocampus Bernadett Boda, Stefano Alberi, Irina Nikonenko, Roxanne Node-Langlois, Pascal Jourdain, Marlyse Moosmayer, Lorena Parisi-Jourdain, and Dominique Muller Department of Basic Neuroscience, Centre Medical Universitaire, 1211 Geneva 4, Switzerland Mutations of the gene coding for PAK3 (p21-activated kinase 3) are associated with X-linked, nonsyndromic forms of mental retardation (MRX) in which the only distinctive clinical feature is the cognitive deficit. The mechanisms through which PAK3 mutation produces the mental handicap remain unclear, although an involvement in the mechanisms that regulate the formation or plasticity of synaptic networks has been proposed. Here we show, using a transient transfection approach, that antisense and small interfering RNA-mediated suppression of PAK3 or expression of a dominant-negative PAK3 carrying the human MRX30 mutation in rat hippocampal organotypic slice cultures results in the formation of abnormally elongated dendritic spines and filopodia-like protrusions and a decrease in mature spine synapses. Ultrastructural analysis of the changes induced by expression of PAK3 carrying the MRX30 mutation reveals that many elongated spines fail to express postsynaptic densities or contact presynaptic terminals. These defects are associated with a reduced spontaneous activity, altered expression of AMPA-type glutamate receptors, and defective long-term potentiation. Together, these data identify PAK3 as a key regulator of synapse formation and plasticity in the hippocampus and support interpretations that these defects might contribute to the cognitive deficits underlying this form of mental retardation. -

A De Novo Mutation in the X-Linked PAK3 Gene Is the Underlying Cause of Intellectual Disability and Macrocephaly in Monozygotic Twins
European Journal of Medical Genetics xxx (2017) 1e5 Contents lists available at ScienceDirect European Journal of Medical Genetics journal homepage: http://www.elsevier.com/locate/ejmg A de novo mutation in the X-linked PAK3 gene is the underlying cause of intellectual disability and macrocephaly in monozygotic twins Jozef Hertecant a, b, Makanko Komara c, Aslam Nagi a, Olfat Al-Zaabi d, Waseem Fathallah e, Hong Cui f, Yaping Yang f, Christine M. Eng f, Mohammad Al Sorkhy g, * Mohammad A. Ghattas g, Lihadh Al-Gazali b, Bassam R. Ali c, a Department of Paediatrics, Tawam Hospital, Al-Ain, United Arab Emirates b Department of Paediatrics, College of Medicine and Health Sciences, United Arab Emirates University, Al-Ain, United Arab Emirates c Department of Pathology, College of Medicine and Health Sciences, United Arab Emirates University, Al-Ain, United Arab Emirates d Fujairah Hospital, Fujairah, United Arab Emirates e Mafraq Hospital, Abu Dhabi, United Arab Emirates f Department of Molecular and Human Genetics, Baylor College of Medicine, Baylor Miraca Genetics Laboratories, Houston, TX 77030, USA g College of Pharmacy, Al Ain University of Science and Technology, Al-Ain, United Arab Emirates article info abstract Article history: Pathogenic variants in theP21 protein (Cdc42/Rac)-activated kinase 3gene (PAK3) lead to a rare non Received 29 May 2016 syndromic X-linked intellectual disability. The protein encoded by this gene forms an activated complex Received in revised form with GTP-bound RAS-like (P21), CDC2 and RAC1 proteins which then mediates a variety of cellular 17 January 2017 processes. So far, mutations in PAK3 gene have been reported in few families affected with intellectual Accepted 18 January 2017 disability associated with neurological manifestations such as speech defect, behavioral problem, brain Available online xxx structural abnormalities, microcephaly and cerebral palsy. -

Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling
International Journal of Molecular Sciences Review Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling Maria Carmela Annunziata 1, Melania Parisi 1, Gabriella Esposito 2 , Gabriella Fabbrocini 1, Rosario Ammendola 2 and Fabio Cattaneo 2,* 1 Department of Clinical Medicine and Surgery, School of Medicine, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (M.C.A.); [email protected] (M.P.); [email protected] (G.F.) 2 Department of Molecular Medicine and Medical Biotechnology, School of Medicine,, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (G.E.); [email protected] (R.A.) * Correspondence: [email protected]; Fax: +39-081-7464-359 Received: 5 May 2020; Accepted: 25 May 2020; Published: 27 May 2020 Abstract: FPR1, FPR2, and FPR3 are members of Formyl Peptides Receptors (FPRs) family belonging to the GPCR superfamily. FPR2 is a low affinity receptor for formyl peptides and it is considered the most promiscuous member of this family. Intracellular signaling cascades triggered by FPRs include the activation of different protein kinases and phosphatase, as well as tyrosine kinase receptors transactivation. Protein kinases and phosphatases act coordinately and any impairment of their activation or regulation represents one of the most common causes of several human diseases. Several phospho-sites has been identified in protein kinases and phosphatases, whose role may be to expand the repertoire of molecular mechanisms of regulation or may be necessary for fine-tuning of switch properties. We previously performed a phospho-proteomic analysis in FPR2-stimulated cells that revealed, among other things, not yet identified phospho-sites on six protein kinases and one protein phosphatase.