Characterization of the Macrophage Transcriptome in Glomerulonephritis-Susceptible and -Resistant Rat Strains
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dynamin Functions and Ligands: Classical Mechanisms Behind
1521-0111/91/2/123–134$25.00 http://dx.doi.org/10.1124/mol.116.105064 MOLECULAR PHARMACOLOGY Mol Pharmacol 91:123–134, February 2017 Copyright ª 2017 by The American Society for Pharmacology and Experimental Therapeutics MINIREVIEW Dynamin Functions and Ligands: Classical Mechanisms Behind Mahaveer Singh, Hemant R. Jadhav, and Tanya Bhatt Department of Pharmacy, Birla Institute of Technology and Sciences Pilani, Pilani Campus, Rajasthan, India Received May 5, 2016; accepted November 17, 2016 Downloaded from ABSTRACT Dynamin is a GTPase that plays a vital role in clathrin-dependent pathophysiology of various disorders, such as Alzheimer’s disease, endocytosis and other vesicular trafficking processes by acting Parkinson’s disease, Huntington’s disease, Charcot-Marie-Tooth as a pair of molecular scissors for newly formed vesicles originating disease, heart failure, schizophrenia, epilepsy, cancer, dominant ’ from the plasma membrane. Dynamins and related proteins are optic atrophy, osteoporosis, and Down s syndrome. This review is molpharm.aspetjournals.org important components for the cleavage of clathrin-coated vesicles, an attempt to illustrate the dynamin-related mechanisms involved phagosomes, and mitochondria. These proteins help in organelle in the above-mentioned disorders and to help medicinal chemists division, viral resistance, and mitochondrial fusion/fission. Dys- to design novel dynamin ligands, which could be useful in the function and mutations in dynamin have been implicated in the treatment of dynamin-related disorders. Introduction GTP hydrolysis–dependent conformational change of GTPase dynamin assists in membrane fission, leading to the generation Dynamins were originally discovered in the brain and identi- of endocytic vesicles (Praefcke and McMahon, 2004; Ferguson at ASPET Journals on September 23, 2021 fied as microtubule binding partners. -

Genome-Wide Analysis of 5-Hmc in the Peripheral Blood of Systemic Lupus Erythematosus Patients Using an Hmedip-Chip
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 35: 1467-1479, 2015 Genome-wide analysis of 5-hmC in the peripheral blood of systemic lupus erythematosus patients using an hMeDIP-chip WEIGUO SUI1*, QIUPEI TAN1*, MING YANG1, QIANG YAN1, HUA LIN1, MINGLIN OU1, WEN XUE1, JIEJING CHEN1, TONGXIANG ZOU1, HUANYUN JING1, LI GUO1, CUIHUI CAO1, YUFENG SUN1, ZHENZHEN CUI1 and YONG DAI2 1Guangxi Key Laboratory of Metabolic Diseases Research, Central Laboratory of Guilin 181st Hospital, Guilin, Guangxi 541002; 2Clinical Medical Research Center, the Second Clinical Medical College of Jinan University (Shenzhen People's Hospital), Shenzhen, Guangdong 518020, P.R. China Received July 9, 2014; Accepted February 27, 2015 DOI: 10.3892/ijmm.2015.2149 Abstract. Systemic lupus erythematosus (SLE) is a chronic, Introduction potentially fatal systemic autoimmune disease characterized by the production of autoantibodies against a wide range Systemic lupus erythematosus (SLE) is a typical systemic auto- of self-antigens. To investigate the role of the 5-hmC DNA immune disease, involving diffuse connective tissues (1) and modification with regard to the onset of SLE, we compared is characterized by immune inflammation. SLE has a complex the levels 5-hmC between SLE patients and normal controls. pathogenesis (2), involving genetic, immunologic and envi- Whole blood was obtained from patients, and genomic DNA ronmental factors. Thus, it may result in damage to multiple was extracted. Using the hMeDIP-chip analysis and valida- tissues and organs, especially the kidneys (3). SLE arises from tion by quantitative RT-PCR (RT-qPCR), we identified the a combination of heritable and environmental influences. differentially hydroxymethylated regions that are associated Epigenetics, the study of changes in gene expression with SLE. -

The Rise and Fall of the Bovine Corpus Luteum
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-6-2017 The Rise and Fall of the Bovine Corpus Luteum Heather Talbott University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Biochemistry Commons, Molecular Biology Commons, and the Obstetrics and Gynecology Commons Recommended Citation Talbott, Heather, "The Rise and Fall of the Bovine Corpus Luteum" (2017). Theses & Dissertations. 207. https://digitalcommons.unmc.edu/etd/207 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. THE RISE AND FALL OF THE BOVINE CORPUS LUTEUM by Heather Talbott A DISSERTATION Presented to the Faculty of the University of Nebraska Graduate College in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biochemistry and Molecular Biology Graduate Program Under the Supervision of Professor John S. Davis University of Nebraska Medical Center Omaha, Nebraska May, 2017 Supervisory Committee: Carol A. Casey, Ph.D. Andrea S. Cupp, Ph.D. Parmender P. Mehta, Ph.D. Justin L. Mott, Ph.D. i ACKNOWLEDGEMENTS This dissertation was supported by the Agriculture and Food Research Initiative from the USDA National Institute of Food and Agriculture (NIFA) Pre-doctoral award; University of Nebraska Medical Center Graduate Student Assistantship; University of Nebraska Medical Center Exceptional Incoming Graduate Student Award; the VA Nebraska-Western Iowa Health Care System Department of Veterans Affairs; and The Olson Center for Women’s Health, Department of Obstetrics and Gynecology, Nebraska Medical Center. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Stelios Pavlidis3, Matthew Loza3, Fred Baribaud3, Anthony
Supplementary Data Th2 and non-Th2 molecular phenotypes of asthma using sputum transcriptomics in UBIOPRED Chih-Hsi Scott Kuo1.2, Stelios Pavlidis3, Matthew Loza3, Fred Baribaud3, Anthony Rowe3, Iaonnis Pandis2, Ana Sousa4, Julie Corfield5, Ratko Djukanovic6, Rene 7 7 8 2 1† Lutter , Peter J. Sterk , Charles Auffray , Yike Guo , Ian M. Adcock & Kian Fan 1†* # Chung on behalf of the U-BIOPRED consortium project team 1Airways Disease, National Heart & Lung Institute, Imperial College London, & Biomedical Research Unit, Biomedical Research Unit, Royal Brompton & Harefield NHS Trust, London, United Kingdom; 2Department of Computing & Data Science Institute, Imperial College London, United Kingdom; 3Janssen Research and Development, High Wycombe, Buckinghamshire, United Kingdom; 4Respiratory Therapeutic Unit, GSK, Stockley Park, United Kingdom; 5AstraZeneca R&D Molndal, Sweden and Areteva R&D, Nottingham, United Kingdom; 6Faculty of Medicine, Southampton University, Southampton, United Kingdom; 7Faculty of Medicine, University of Amsterdam, Amsterdam, Netherlands; 8European Institute for Systems Biology and Medicine, CNRS-ENS-UCBL, Université de Lyon, France. †Contributed equally #Consortium project team members are listed under Supplementary 1 Materials *To whom correspondence should be addressed: [email protected] 2 List of the U-BIOPRED Consortium project team members Uruj Hoda & Christos Rossios, Airways Disease, National Heart & Lung Institute, Imperial College London, UK & Biomedical Research Unit, Biomedical Research Unit, Royal -

Discovery of the Novel Autophagy Inhibitor Aumitin That Targets Mitochondrial Complex I
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2018 Discovery of the novel autophagy inhibitor Aumitin that targets mitochondrial complex I Lucas Robkea,b,c, Yushi Futamurad, Georgios Konstantinidise, Julian Wilkea,b, Harumi Aonod, Zhwan Mahmoudb, Nobumoto Watanabec,f, Yao-Wen Wue, Hiroyuki Osadac,d, Luca Laraiaa,g *, Herbert Waldmanna,b * a: Max-Planck-Institute of Molecular Physiology, department of Chemical Biology, Otto-Hahn-Str. 11, 44227 Dortmund (Germany); b: Faculty of Chemistry and Chemical Biology, TU Dortmund University, Otto-Hahn-Str. 4a, 44227 Dortmund (Germany); c: RIKEN-Max Planck Joint Research Division for Systems Chemical Biology, RIKEN CSRS, 2-1, Hirosawa, Wako, Saitama 351-0198 (Japan); d: Chemical Biology Research Group, RIKEN CSRS, 2-1, Hirosawa, Wako, Saitama 351-0198 (Japan); e: Chemical Genomics Centre of the Max-Planck-Society, Otto- Hahn-Str. 15, 44227 Dortmund (Germany); f: Bio-Active Compounds Discovery Research Unit, RIKEN CSRS, 2-1, Hirosawa, Wako, Saitama 351-0198 (Japan). g: present address: Department of Chemistry, Technical University of Denmark, Kemitorvet Building 207, Room 124, 2800 Kgs. Lyngby, Denmark. * [email protected], [email protected] SI-Table 1: Structure activity relationship of the di-aminopyrimidines. Starvation = starvation induced autophagy assay; Rapamycin = Rapamycin induced autophagy assay; Viability = survival assessed by means of an ADP-glow assay. > 10 = no inhibition at a test concentration of 10 -

Supplementary Table 3 Complete List of RNA-Sequencing Analysis of Gene Expression Changed by ≥ Tenfold Between Xenograft and Cells Cultured in 10%O2
Supplementary Table 3 Complete list of RNA-Sequencing analysis of gene expression changed by ≥ tenfold between xenograft and cells cultured in 10%O2 Expr Log2 Ratio Symbol Entrez Gene Name (culture/xenograft) -7.182 PGM5 phosphoglucomutase 5 -6.883 GPBAR1 G protein-coupled bile acid receptor 1 -6.683 CPVL carboxypeptidase, vitellogenic like -6.398 MTMR9LP myotubularin related protein 9-like, pseudogene -6.131 SCN7A sodium voltage-gated channel alpha subunit 7 -6.115 POPDC2 popeye domain containing 2 -6.014 LGI1 leucine rich glioma inactivated 1 -5.86 SCN1A sodium voltage-gated channel alpha subunit 1 -5.713 C6 complement C6 -5.365 ANGPTL1 angiopoietin like 1 -5.327 TNN tenascin N -5.228 DHRS2 dehydrogenase/reductase 2 leucine rich repeat and fibronectin type III domain -5.115 LRFN2 containing 2 -5.076 FOXO6 forkhead box O6 -5.035 ETNPPL ethanolamine-phosphate phospho-lyase -4.993 MYO15A myosin XVA -4.972 IGF1 insulin like growth factor 1 -4.956 DLG2 discs large MAGUK scaffold protein 2 -4.86 SCML4 sex comb on midleg like 4 (Drosophila) Src homology 2 domain containing transforming -4.816 SHD protein D -4.764 PLP1 proteolipid protein 1 -4.764 TSPAN32 tetraspanin 32 -4.713 N4BP3 NEDD4 binding protein 3 -4.705 MYOC myocilin -4.646 CLEC3B C-type lectin domain family 3 member B -4.646 C7 complement C7 -4.62 TGM2 transglutaminase 2 -4.562 COL9A1 collagen type IX alpha 1 chain -4.55 SOSTDC1 sclerostin domain containing 1 -4.55 OGN osteoglycin -4.505 DAPL1 death associated protein like 1 -4.491 C10orf105 chromosome 10 open reading frame 105 -4.491 -

PI3K Catalytic Isoform Alteration Promotes the LIMK1-Related
ANTICANCER RESEARCH 37 : 1805-1818 (2017) doi:10.21873/anticanres.11515 PI3K Catalytic Isoform Alteration Promotes the LIMK1-related Metastasis Through the PAK1 or ROCK1/2 Activation in Cigarette Smoke-exposed Ovarian Cancer Cells GA BIN PARK 1 and DAEJIN KIM 2 1Department of Biochemistry, Kosin University College of Medicine, Busan, Republic of Korea; 2Department of Anatomy, Inje University College of Medicine, Busan, Republic of Korea Abstract. Aim: To investigate the molecular mechanisms Several studies have shown a strong correlation between by which long-term exposure to cigarette smoke extract cigarette smoke (CS) and cancer metastasis through the (CSE) contributes to ovarian cancer metastasis. Materials induction of numerous factors involved in migration activity and Methods: Western blot analysis for diverse p110 (1-3). The exposure to CS induces the epithelial- isoforms of phosphoinositide 3-kinase (PI3K)-related mesenchymal transition (EMT) process and up-regulates the signaling pathway and epithelial-mesenchymal transition expression of EMT markers, including N-cadherin and (EMT) markers was performed to analyze the underlying vimentin (4, 5). Cigarette smoke extract (CSE) treatment mechanisms. Migratory activity of CSE-exposed ovarian significantly induces interleukin-8 (IL-8) and transforming cancer cells was determined by transendothelial migration growth factor-beta 1 (TGF- β1 ) production and profoundly and invasion assay. Results: After exposure to CSE for four suppresses the proliferation and growth of erythroid and weeks, CaOV3 (primary) and SKOV3 (metastatic) ovarian granulocyte-macrophage progenitors (6). Stimulation with cancer cells showed enhanced mesenchymal characteristics CSE in human lung fibroblast cells induces the expression and produced EMT-related cytokines [intwerleukin-8 (IL-8), of phosphorylated Smad3, a main downstream target of the vascular endothelial growth factor (VEGF) and TGF- β1 receptor, which results in the secretion of vascular transforming growth factor-beta 1 (TGF- β1 )]. -

Circular RNA Hsa Circ 0005114‑Mir‑142‑3P/Mir‑590‑5P‑ Adenomatous
ONCOLOGY LETTERS 21: 58, 2021 Circular RNA hsa_circ_0005114‑miR‑142‑3p/miR‑590‑5p‑ adenomatous polyposis coli protein axis as a potential target for treatment of glioma BO WEI1*, LE WANG2* and JINGWEI ZHAO1 1Department of Neurosurgery, China‑Japan Union Hospital of Jilin University, Changchun, Jilin 130033; 2Department of Ophthalmology, The First Hospital of Jilin University, Jilin University, Changchun, Jilin 130021, P.R. China Received September 12, 2019; Accepted October 22, 2020 DOI: 10.3892/ol.2020.12320 Abstract. Glioma is the most common type of brain tumor APC expression with a good overall survival rate. UALCAN and is associated with a high mortality rate. Despite recent analysis using TCGA data of glioblastoma multiforme and the advances in treatment options, the overall prognosis in patients GSE25632 and GSE103229 microarray datasets showed that with glioma remains poor. Studies have suggested that circular hsa‑miR‑142‑3p/hsa‑miR‑590‑5p was upregulated and APC (circ)RNAs serve important roles in the development and was downregulated. Thus, hsa‑miR‑142‑3p/hsa‑miR‑590‑5p‑ progression of glioma and may have potential as therapeutic APC‑related circ/ceRNA axes may be important in glioma, targets. However, the expression profiles of circRNAs and their and hsa_circ_0005114 interacted with both of these miRNAs. functions in glioma have rarely been studied. The present study Functional analysis showed that hsa_circ_0005114 was aimed to screen differentially expressed circRNAs (DECs) involved in insulin secretion, while APC was associated with between glioma and normal brain tissues using sequencing the Wnt signaling pathway. In conclusion, hsa_circ_0005114‑ data collected from the Gene Expression Omnibus database miR‑142‑3p/miR‑590‑5p‑APC ceRNA axes may be potential (GSE86202 and GSE92322 datasets) and explain their mecha‑ targets for the treatment of glioma. -
![Downloaded from [266]](https://docslib.b-cdn.net/cover/7352/downloaded-from-266-347352.webp)
Downloaded from [266]
Patterns of DNA methylation on the human X chromosome and use in analyzing X-chromosome inactivation by Allison Marie Cotton B.Sc., The University of Guelph, 2005 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in The Faculty of Graduate Studies (Medical Genetics) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2012 © Allison Marie Cotton, 2012 Abstract The process of X-chromosome inactivation achieves dosage compensation between mammalian males and females. In females one X chromosome is transcriptionally silenced through a variety of epigenetic modifications including DNA methylation. Most X-linked genes are subject to X-chromosome inactivation and only expressed from the active X chromosome. On the inactive X chromosome, the CpG island promoters of genes subject to X-chromosome inactivation are methylated in their promoter regions, while genes which escape from X- chromosome inactivation have unmethylated CpG island promoters on both the active and inactive X chromosomes. The first objective of this thesis was to determine if the DNA methylation of CpG island promoters could be used to accurately predict X chromosome inactivation status. The second objective was to use DNA methylation to predict X-chromosome inactivation status in a variety of tissues. A comparison of blood, muscle, kidney and neural tissues revealed tissue-specific X-chromosome inactivation, in which 12% of genes escaped from X-chromosome inactivation in some, but not all, tissues. X-linked DNA methylation analysis of placental tissues predicted four times higher escape from X-chromosome inactivation than in any other tissue. Despite the hypomethylation of repetitive elements on both the X chromosome and the autosomes, no changes were detected in the frequency or intensity of placental Cot-1 holes. -
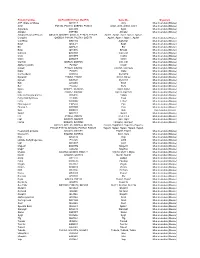
Gene Ids Organism ATP Citrate Synthase Q3V117 Acly
Protein Families UniProtKB ID (from MudPIT) Gene IDs Organism ATP citrate synthase Q3V117 Acly Mus musculus (Mouse) Actin P68134, P60710, Q8BFZ3, P68033 Acta1, Actb, Actbl2, Actc1 Mus musculus (Mouse) Argonaute Q8CJG0 Ago2 Mus musculus (Mouse) Ahnak2 E9PYB0 Ahnak2 Mus musculus (Mouse) Adaptor Related Protein Q8CC13, Q8CBB7, Q3UHJ0, P17426, P17427, Ap1b1, Ap1g1, Aak1, Ap2a1, Ap2a2, Complex Q9DBG3, P84091, P62743, Q9Z1T1 Ap2b1, Ap2m1, Ap2s1 , Ap3b1 Mus musculus (Mouse) V-ATPase Q9Z1G4 Atp6v0a1 Mus musculus (Mouse) Bag3 Q9JLV1 Bag3 Mus musculus (Mouse) Bcr Q6PAJ1 Bcr Mus musculus (Mouse) Bmp Q91Z96 Bmp2k Mus musculus (Mouse) Calcoco Q8CGU1 Calcoco1 Mus musculus (Mouse) Ccdc D3YZP9 Ccdc6 Mus musculus (Mouse) Clint1 Q5SUH7 Clint1 Mus musculus (Mouse) Clathrin Q6IRU5, Q68FD5 Cltb, Cltc Mus musculus (Mouse) Alpha-crystallin P23927 Cryab Mus musculus (Mouse) Casein P19228, Q02862 Csn1s1, Csn1s2a Mus musculus (Mouse) Dab2 P98078 Dab2 Mus musculus (Mouse) Connecdenn Q8K382 Dennd1a Mus musculus (Mouse) Dynamin P39053, P39054 Dnm1, Dnm2 Mus musculus (Mouse) Dynein Q9JHU4 Dync1h1 Mus musculus (Mouse) Edc Q3UJB9 Edc4 Mus musculus (Mouse) Eef P58252 Eef2 Mus musculus (Mouse) Epsin Q80VP1, Q5NCM5 Epn1, Epn2 Mus musculus (Mouse) Eps P42567, Q60902 Eps15, Eps15l1 Mus musculus (Mouse) Fatty acid binding protein Q05816 Fabp5 Mus musculus (Mouse) Fatty Acid Synthase P19096 Fasn Mus musculus (Mouse) Fcho Q3UQN2 Fcho2 Mus musculus (Mouse) Fibrinogen A E9PV24 Fga Mus musculus (Mouse) Filamin A Q8BTM8 Flna Mus musculus (Mouse) Gak Q99KY4 Gak Mus musculus (Mouse) -

Role of RUNX1 in Aberrant Retinal Angiogenesis Jonathan D
Page 1 of 25 Diabetes Identification of RUNX1 as a mediator of aberrant retinal angiogenesis Short Title: Role of RUNX1 in aberrant retinal angiogenesis Jonathan D. Lam,†1 Daniel J. Oh,†1 Lindsay L. Wong,1 Dhanesh Amarnani,1 Cindy Park- Windhol,1 Angie V. Sanchez,1 Jonathan Cardona-Velez,1,2 Declan McGuone,3 Anat O. Stemmer- Rachamimov,3 Dean Eliott,4 Diane R. Bielenberg,5 Tave van Zyl,4 Lishuang Shen,1 Xiaowu Gai,6 Patricia A. D’Amore*,1,7 Leo A. Kim*,1,4 Joseph F. Arboleda-Velasquez*1 Author affiliations: 1Schepens Eye Research Institute/Massachusetts Eye and Ear, Department of Ophthalmology, Harvard Medical School, 20 Staniford St., Boston, MA 02114 2Universidad Pontificia Bolivariana, Medellin, Colombia, #68- a, Cq. 1 #68305, Medellín, Antioquia, Colombia 3C.S. Kubik Laboratory for Neuropathology, Massachusetts General Hospital, 55 Fruit St., Boston, MA 02114 4Retina Service, Massachusetts Eye and Ear Infirmary, Department of Ophthalmology, Harvard Medical School, 243 Charles St., Boston, MA 02114 5Vascular Biology Program, Boston Children’s Hospital, Department of Surgery, Harvard Medical School, 300 Longwood Ave., Boston, MA 02115 6Center for Personalized Medicine, Children’s Hospital Los Angeles, Los Angeles, 4650 Sunset Blvd, Los Angeles, CA 90027, USA 7Department of Pathology, Harvard Medical School, 25 Shattuck St., Boston, MA 02115 Corresponding authors: Joseph F. Arboleda-Velasquez: [email protected] Ph: (617) 912-2517 Leo Kim: [email protected] Ph: (617) 912-2562 Patricia D’Amore: [email protected] Ph: (617) 912-2559 Fax: (617) 912-0128 20 Staniford St. Boston MA, 02114 † These authors contributed equally to this manuscript Word Count: 1905 Tables and Figures: 4 Diabetes Publish Ahead of Print, published online April 11, 2017 Diabetes Page 2 of 25 Abstract Proliferative diabetic retinopathy (PDR) is a common cause of blindness in the developed world’s working adult population, and affects those with type 1 and type 2 diabetes mellitus.