Phosphoproteomic Analysis of Lung Tissue in Pahreveals Activation Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MAP2K4/MKK4 Expression in Pancreatic Cancer: Genetic Validation of Immunohistochemistry and Relationship to Disease Course
8516 Vol. 10, 8516–8520, December 15, 2004 Clinical Cancer Research MAP2K4/MKK4 Expression in Pancreatic Cancer: Genetic Validation of Immunohistochemistry and Relationship to Disease Course Wei Xin,1 Ki J. Yun,4 Francesca Ricci,2 ing patterns were also evaluated among unresectable pri- Marianna Zahurak,3 Wanglong Qiu,4 mary and metastatic cancer tissues from autopsy specimens, Gloria H. Su,4 Charles J. Yeo,5,6 indicating intact Mkk4 immunolabeling in 88.8% of the unresectable primary carcinomas as compared with 63.3% Ralph H. Hruban,4,5 Scott E. Kern,4,5 and 4,5 of distant metastases (P < 0.001). Our data indicate that the Christine A. Iacobuzio-Donahue loss of Mkk4 protein expression in pancreatic carcinomas 1 Department of Pathology, The University of Michigan Medical may be more frequent than suggested by the rates of genetic Center, Ann Arbor, Michigan; 2Department of Pathology, University La Sapienza, Rome, Italy; and the Departments of 3Biostatistics, inactivation alone and that MKK4 loss may contribute to 4Pathology, 5Oncology, and 6Surgery, The Johns Hopkins University disease progression. The correlation of MKK4 genetic status Hospital, Baltimore, Maryland with immunolabeling patterns validate this approach for the evaluation of MKK4 status in routine histologic sections and ABSTRACT may provide useful information regarding patient prognosis. MKK4 (MAP2K4/SEK1) is a member of the mitogen- activated protein kinase family, originally identified as a INTRODUCTION kinase involved in the stress-activated protein kinase path- The mitogen-activated protein (MAP) kinase cascades are way by directly phosphorylating c-Jun NH -terminal kinase. 2 multifunctional signaling pathways that are evolutionally well MKK4 genetic inactivation has been observed in a subset of conserved in all of the eukaryotic cells. -
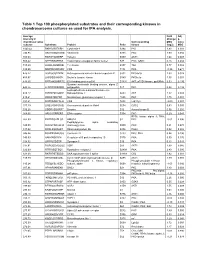
Table 1 Top 100 Phosphorylated Substrates and Their Corresponding Kinases in Chondrosarcoma Cultures As Used for IPA Analysis
Table 1 Top 100 phosphorylated substrates and their corresponding kinases in chondrosarcoma cultures as used for IPA analysis. Average Fold Adj intensity in Change p- chondrosarcoma Corresponding MSC value cultures Substrate Protein Psite kinase (log2) MSC 1043.42 RKKKVSSTKRH Cytohesin-1 S394 PKC 1.83 0.001 746.95 RKGYRSQRGHS Vitronectin S381 PKC 1.00 0.056 709.03 RARSTSLNERP Tuberin S939 AKT1 1.64 0.008 559.42 SPPRSSLRRSS Transcription elongation factor A-like1 S37 PKC; GSK3 0.18 0.684 515.29 LRRSLSRSMSQ Telethonin S157 Titin 0.77 0.082 510.00 MQPDNSSDSDY CD5 T434 PKA -0.35 0.671 476.27 GGRGGSRARNL Heterogeneous nuclear ribonucleoprotein K S302 PKCdelta 1.03 0.028 455.97 LKPGSSHRKTK Bruton's tyrosine kinase S180 PKCbeta 1.55 0.001 444.65 RRRMASMQRTG E1A binding protein p300 S1834 AKT; p70S6 kinase; pp90Rsk 0.53 0.195 Guanine nucleotide binding protein, alpha Z 440.26 HLRSESQRQRR polypeptide S27 PKC 0.88 0.199 6-phosphofructo-2-kinase/fructose-2,6- 424.12 RPRNYSVGSRP biphosphatase 2 S483 AKT 1.32 0.003 419.61 KKKIATRKPRF Metabotropic glutamate receptor 1 T695 PKC 1.75 0.001 391.21 DNSSDSDYDLH CD5 T453 Lck; Fyn -2.09 0.001 377.39 LRQLRSPRRAQ Ras associated protein Rab4 S204 CDC2 0.63 0.091 376.28 SSQRVSSYRRT Desmin S12 Aurora kinase B 0.56 0.255 369.05 ARIGGSRRERS EP4 receptor S354 PKC 0.29 0.543 RPS6 kinase alpha 3; PKA; 367.99 EPKRRSARLSA HMG14 S7 PKC -0.01 0.996 Peptidylglycine alpha amidating 349.08 SRKGYSRKGFD monooxygenase S930 PKC 0.21 0.678 347.92 RRRLSSLRAST Ribosomal protein S6 S236 PAK2 0.02 0.985 346.84 RSNPPSRKGSG Connexin -

Protein-Protein Interactions Among Signaling Pathways May Become New Therapeutic Targets in Liver Cancer (Review)
ONCOLOGY REPORTS 35: 625-638, 2016 Protein-protein interactions among signaling pathways may become new therapeutic targets in liver cancer (Review) XIAO ZHANG1*, YULAN WANG1*, Jiayi WANG1,2 and FENYONG SUN1 1Department of Clinical Laboratory Medicine, Shanghai Tenth People's Hospital of Tongji University, Shanghai 200072; 2Translation Medicine of High Institute, Tongji University, Shanghai 200092, P.R. China Received May 29, 2015; Accepted July 6, 2015 DOI: 10.3892/or.2015.4464 Abstract. Numerous signaling pathways have been shown to be 1. Introduction dysregulated in liver cancer. In addition, some protein-protein interactions are prerequisite for the uncontrolled activation Liver cancer is the sixth most common cancer and the second or inhibition of these signaling pathways. For instance, in most common cause of cancer-associated mortality world- the PI3K/AKT signaling pathway, protein AKT binds with wide (1). Approximately 75% of all primary liver cancer types a number of proteins such as mTOR, FOXO1 and MDM2 to are hepatocellular carcinoma (HCC) that formed from liver play an oncogenic role in liver cancer. The aim of the present cells. Liver cancer can be formed from other structures in review was to focus on a series of important protein-protein the liver such as bile duct, blood vessels and immune cells. interactions that can serve as potential therapeutic targets Secondary liver cancer is a result of metastasis of cancer from in liver cancer among certain important pro-carcinogenic other body sites into the liver. The major cause of primary liver signaling pathways. The strategies of how to investigate and cancer is viral infection with either hepatitis C virus (HCV) analyze the protein-protein interactions are also included in or hepatitis B virus (HBV), which leads to massive inflamma- this review. -

Inhibition of ERK 1/2 Kinases Prevents Tendon Matrix Breakdown Ulrich Blache1,2,3, Stefania L
www.nature.com/scientificreports OPEN Inhibition of ERK 1/2 kinases prevents tendon matrix breakdown Ulrich Blache1,2,3, Stefania L. Wunderli1,2,3, Amro A. Hussien1,2, Tino Stauber1,2, Gabriel Flückiger1,2, Maja Bollhalder1,2, Barbara Niederöst1,2, Sandro F. Fucentese1 & Jess G. Snedeker1,2* Tendon extracellular matrix (ECM) mechanical unloading results in tissue degradation and breakdown, with niche-dependent cellular stress directing proteolytic degradation of tendon. Here, we show that the extracellular-signal regulated kinase (ERK) pathway is central in tendon degradation of load-deprived tissue explants. We show that ERK 1/2 are highly phosphorylated in mechanically unloaded tendon fascicles in a vascular niche-dependent manner. Pharmacological inhibition of ERK 1/2 abolishes the induction of ECM catabolic gene expression (MMPs) and fully prevents loss of mechanical properties. Moreover, ERK 1/2 inhibition in unloaded tendon fascicles suppresses features of pathological tissue remodeling such as collagen type 3 matrix switch and the induction of the pro-fbrotic cytokine interleukin 11. This work demonstrates ERK signaling as a central checkpoint to trigger tendon matrix degradation and remodeling using load-deprived tissue explants. Tendon is a musculoskeletal tissue that transmits muscle force to bone. To accomplish its biomechanical function, tendon tissues adopt a specialized extracellular matrix (ECM) structure1. Te load-bearing tendon compart- ment consists of highly aligned collagen-rich fascicles that are interspersed with tendon stromal cells. Tendon is a mechanosensitive tissue whereby physiological mechanical loading is vital for maintaining tendon archi- tecture and homeostasis2. Mechanical unloading of the tissue, for instance following tendon rupture or more localized micro trauma, leads to proteolytic breakdown of the tissue with severe deterioration of both structural and mechanical properties3–5. -

PRODUCTS and SERVICES Target List
PRODUCTS AND SERVICES Target list Kinase Products P.1-11 Kinase Products Biochemical Assays P.12 "QuickScout Screening Assist™ Kits" Kinase Protein Assay Kits P.13 "QuickScout Custom Profiling & Panel Profiling Series" Targets P.14 "QuickScout Custom Profiling Series" Preincubation Targets Cell-Based Assays P.15 NanoBRET™ TE Intracellular Kinase Cell-Based Assay Service Targets P.16 Tyrosine Kinase Ba/F3 Cell-Based Assay Service Targets P.17 Kinase HEK293 Cell-Based Assay Service ~ClariCELL™ ~ Targets P.18 Detection of Protein-Protein Interactions ~ProbeX™~ Stable Cell Lines Crystallization Services P.19 FastLane™ Structures ~Premium~ P.20-21 FastLane™ Structures ~Standard~ Kinase Products For details of products, please see "PRODUCTS AND SERVICES" on page 1~3. Tyrosine Kinases Note: Please contact us for availability or further information. Information may be changed without notice. Expression Protein Kinase Tag Carna Product Name Catalog No. Construct Sequence Accession Number Tag Location System HIS ABL(ABL1) 08-001 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) BTN BTN-ABL(ABL1) 08-401-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ABL(ABL1) [E255K] HIS ABL(ABL1)[E255K] 08-094 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) HIS ABL(ABL1)[T315I] 08-093 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) [T315I] BTN BTN-ABL(ABL1)[T315I] 08-493-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ACK(TNK2) GST ACK(TNK2) 08-196 Catalytic domain -

An Exploration of Mutation Status of Cancer Genes in Breast Cancers
on: Sequ ati en er c n in e g G & t x A e p Journal of Next Generation p N l f i c o Wang, Next Generat Sequenc & Applic 2014, 1:1 a l t a i o n r ISSN: 2469-9853n u s o DOI: 10.4172/2469-9853.1000103 J Sequencing & Applications Research Article Open Access An Exploration of Mutation Status of Cancer Genes in Breast Cancers Xiaosheng Wang* Department of Genetics, Cell Biology and Anatomy, University of Nebraska Medical Center, Omaha, USA *Corresponding author: Xiaosheng Wang, Department of Genetics, Cell Biology and Anatomy, University of Nebraska Medical Center, Omaha, USA, Tel: 402-559-4813; E-mail: [email protected] Rec date: Jan 21, 2014, Acc date: Apr 26, 2014, Pub date: Apr 28, 2014 Copyright: © 2014 Wang X. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction H2O in any medium, provided the original author and source are credited. Abstract Breast cancer is the most common cancer in women in US, and has the second highest mortality rate that accounts for about 25% of all cancer deaths. It has been recognized that genetic biomarkers for cancer are useful for estimating the cancer recurrence risk, and guiding targeted treatment of cancer. Since breast cancers carry a wide spectrum of gene mutations in their genomes, identification of these mutations would be promising in improving diagnosis and treatment of breast cancers. The rapid advances in Next-Generation Sequencing (NGS) technology have generated a large amount of NGS data on breast cancer genomes that makes detection and application of mutant biomarkers for breast cancer a reality. -

The Use of Genetic Analyses and Functional Assays for the Interpretation of Rare Variants in Pediatric Heart Disease
The use of genetic analyses and functional assays for the interpretation of rare variants in pediatric heart disease A dissertation submitted to the Division of Graduate Studies and Research, University of Cincinnati in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Molecular Genetics by Jeffrey A. Schubert Bachelor of Science, Mount St. Joseph University, 2012 Committee Chair: Stephanie M. Ware, M.D., Ph.D. Edmund Choi, Ph.D. Benjamin Landis, M.D. Anil Menon, Ph.D. David Wieczorek, Ph.D. Molecular Genetics, Biochemistry, and Microbiology Graduate Program College of Medicine, University of Cincinnati Cincinnati, Ohio, USA, 2018 ABSTRACT The use of next generation technologies such as whole exome sequencing (WES) has paved the way for discovering novel causes of Mendelian diseases. This has been demonstrated in pediatric heart diseases, including cardiomyopathy (CM) and familial thoracic aortic aneurysm (TAA). Each of these conditions carries a high risk of a serious cardiac event, including sudden heart failure or aortic rupture, which are often fatal. Patients with either disease can be asymptomatic before presenting with these events, which necessitates early diagnosis. Though there are many known genetic causes of disease for both conditions, there is still room for discovery of novel pathogenic genes and variants, as many patients have an undefined genetic diagnosis. WES covers the protein-coding portion of the genome, which yields a massive amount of data, though it comprises only 1% of the genome. Sorting and filtering sequencing information to identify (sometimes) a single base pair change responsible for the patient phenotype is challenging. Further, interpreting identified candidate variants must be done according to strict standards, which makes it difficult to definitively say whether a coding change is pathogenic or benign. -

WO 2017/083562 Al 18 May 20 17 (18.05.2017) W P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2017/083562 Al 18 May 20 17 (18.05.2017) W P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C12Q 1/68 (2006.01) C12N 15/10 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (21) International Application Number: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, PCT/US2016/061395 KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (22) International Filing Date: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, 10 November 2016 (10.1 1.2016) OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (25) Filing Language: English TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, (26) Publication Language: English ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 62/254,1 10 11 November 2015 ( 11. 11.2015) US kind of regional protection available): ARIPO (BW, GH, GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, (71) Applicant: RESOLUTION BIOSCIENCE, INC. TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, [US/US]; 2023 120th Avenue Northeast, Suite 100, Bel- TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, levue, Washington 98005 (US). -

Targeted MS Quantitation Assays for Signal Transduction Protein Pathways
Targeted MS quantitation assays for signal transduction protein pathways Paul Haney R&D Platform Manager Thermo Scientific Protein Research Products Rockford, IL Why Targeted Quantitative Proteomics via Mass Spec? . Measure protein abundance and protein isoforms (e.g. splice variants, PTMs) without the need for antibodies . Avoid immuno-based cross- reactivity during multiplexing. Validate relative quantitation data from discovery proteomic experiments 2 How Do You Select Peptides for Targeted MS Assays? List of Proteins Spectral library Discovery data repositories In silico prediction Pinpoint 1.2 Hypothesis Experimental List of target peptides and transitions 3 How Do You Select Peptides for Targeted MS Assays? List of Proteins Spectral library Discovery data repositories In silico prediction Pinpoint 1.2 Hypothesis Experimental Validation Assay results List of target peptides and transitions 4 Tools for Target Peptide Identification and Scheduling . Active-site probes for enzyme subclass enrichment . Rapid recombinant heavy protein expression using human cell-free extracts . Peptide retention time calibration mixture for chromatography QC and targeted method 100 80 60 acquisition scheduling Intensity 40 20 0 5 10 15 20 25 30 5 Tools for Target Peptide Identification and Scheduling . Active-site probes for enzyme subclass enrichment . Rapid recombinant heavy protein expression using human cell-free extracts • Stergachis, A. & MacCoss, M. (2011) Nature Methods (submitted) . Peptide retention time calibration mixture for 100 80 chromatography -

Combined Mtorc1/Mtorc2 Inhibition Blocks Growth and Induces Catastrophic Macropinocytosis in Cancer Cells
Combined mTORC1/mTORC2 inhibition blocks growth and induces catastrophic macropinocytosis in cancer cells Ritesh K. Srivastavaa, Changzhao Lia, Jasim Khana, Nilam Sanjib Banerjeeb, Louise T. Chowb,1, and Mohammad Athara,1 aDepartment of Dermatology, University of Alabama at Birmingham, Birmingham, AL 35294; and bDepartment of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, Birmingham, AL 35294 Contributed by Louise T. Chow, October 4, 2019 (sent for review July 3, 2019; reviewed by Hasan Mukhtar and Brian A. Van Tine) The mammalian target of rapamycin (mTOR) pathway, which plays mTORC2 (9). Aberrant activation of these components of mTOR a critical role in regulating cellular growth and metabolism, is signaling pathways is associated with many cancer types, including aberrantly regulated in the pathogenesis of a variety of neoplasms. those that develop in the skin, lung, colon, breast, and brain (10–12). Here we demonstrate that dual mTORC1/mTORC2 inhibitors OSI-027 Recently, we and others have reported an association of mTOR and PP242 cause catastrophic macropinocytosis in rhabdomyosar- up-regulation with rhabdomyosarcoma (RMS) tumor progression coma (RMS) cells and cancers of the skin, breast, lung, and cervix, (13, 14). Although mTORC1 inhibitors initially exhibited some whereas the effects are much less pronounced in immortalized inhibitory effects, the tumors became resistant due to feedback human keratinocytes. Using RMS as a model, we characterize in activation of AKT signaling by the mTORC2-regulated phos- detail the mechanism of macropinocytosis induction. Macropino- phorylation (15). Therefore, extensive efforts are now ongoing to somes are distinct from endocytic vesicles and autophagosomes in develop potent inhibitors that could simultaneously target both that they are single-membrane bound vacuoles formed by projection, mTORC1 and mTORC2 signaling pathways (16, 17). -

Dema and Faust Et Al., Suppl. Material 2020.02.03
Supplementary Materials Cyclin-dependent kinase 18 controls trafficking of aquaporin-2 and its abundance through ubiquitin ligase STUB1, which functions as an AKAP Dema Alessandro1,2¶, Dörte Faust1¶, Katina Lazarow3, Marc Wippich3, Martin Neuenschwander3, Kerstin Zühlke1, Andrea Geelhaar1, Tamara Pallien1, Eileen Hallscheidt1, Jenny Eichhorst3, Burkhard Wiesner3, Hana Černecká1, Oliver Popp1, Philipp Mertins1, Gunnar Dittmar1, Jens Peter von Kries3, Enno Klussmann1,4* ¶These authors contributed equally to this work 1Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC), Robert- Rössle-Strasse 10, 13125 Berlin, Germany 2current address: University of California, San Francisco, 513 Parnassus Avenue, CA 94122 USA 3Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Robert-Rössle-Strasse 10, 13125 Berlin, Germany 4DZHK (German Centre for Cardiovascular Research), Partner Site Berlin, Oudenarder Strasse 16, 13347 Berlin, Germany *Corresponding author Enno Klussmann Max Delbrück Center for Molecular Medicine Berlin in the Helmholtz Association (MDC) Robert-Rössle-Str. 10, 13125 Berlin Germany Tel. +49-30-9406 2596 FAX +49-30-9406 2593 E-mail: [email protected] 1 Content 1. CELL-BASED SCREENING BY AUTOMATED IMMUNOFLUORESCENCE MICROSCOPY 3 1.1 Screening plates 3 1.2 Image analysis using CellProfiler 17 1.4 Identification of siRNA affecting cell viability 18 1.7 Hits 18 2. SUPPLEMENTARY TABLE S4, FIGURES S2-S4 20 2 1. Cell-based screening by automated immunofluorescence microscopy 1.1 Screening plates Table S1. Genes targeted with the Mouse Protein Kinases siRNA sub-library. Genes are sorted by plate and well. Accessions refer to National Center for Biotechnology Information (NCBI, BLA) entries. The siRNAs were arranged on three 384-well microtitre platres. -

Mutual Exclusivity: Drivers, Pathways, and Beyond
Mutual exclusivity: drivers, pathways, and beyond Teresa Przytycka NIH / NLM / NCBI Cancer drivers, passengers, supporting actors, witnesses • Driver mutations /alterations– mutations contributing to cancer progression • Passenger mutations – neutral mutations accumulating during cancer progression • Challenges in detecting driver mutations: – Heterogeneity - phenotypically similar cancer cases might be caused by different sets of driver mutations – Rare drivers • Best supporting actors (Igor’s talk) • Witnesses (this talk) Cancer driving pathways examples of BRCA mutated genes in their pathway context RAS 1.5% or more PIK3CR1 PTEN PIK3CA CTCF AKT1 MAPK signaling FOXA1 MAP3K1 MAP3K4 SWI/SNF MAP2K4 ARID1A mTOR NCOA3 EP300 CEBPA Mediator complex co –activator /co-repressor MED23 co –activation NCOR1 Mutual exclusivity of cancer drivers Thomas et al 2007 Ciriello, et al., 2012; Vandin, et al., 2012; Szczuret et.al , 2014, 2015 Leiserson, et al., Vadin et al. 2013,2014,2015; Kim et al. 2015 Constantinescu et al. 2015 patients mutations in gene 1 Mutations in gene 2 Explanations • Two drivers dysregulating the same pathway • Each of the drivers corresponds to of a unique cancer type or subtype • Negative genetic interactions between drivers 4 Mutually exclusive pairs often share pathways RAS PIK3CR1 PTEN PIK3CA CTCF AKT1 MAPK signaling FOXA1 MAP3K1 MAP3K4 SWI/SNF MAP2K4 ARID1A mTOR NCOA3 EP300 CEBPA Mediator complex co –activator /co-repressor MED23 co –activation NCOR1 5 Fabio Vandin et al. Genome Res. 2012;22:375-385 Mutually exclusive pairs often share pathways RAS PIK3CR1 PTEN PIK3CA CTCF AKT1 MAPK signaling FOXA1 MAP3K1 MAP3K4 SWI/SNF MAP2K4 ARID1A mTOR NCOA3 EP300 CEBPA Mediator complex co –activator /co-repressor MED23 co –activation NCOR1 6 Mutually exclusive pairs often share pathways RAS PIK3CR1 PTEN PIK3CA CTCF AKT1 MAPK signaling FOXA1 MAP3K1 MAP3K4 SWI/SNF MAP2K4 ARID1A mTOR NCOA3 EP300 CEBPA Mediator complex co –activator /co-repressor MED23 co –activation NCOR1 7 Fabio Vandin et al.