Mutual Exclusivity: Drivers, Pathways, and Beyond
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

AGC Kinases in Mtor Signaling, in Mike Hall and Fuyuhiko Tamanoi: the Enzymes, Vol
Provided for non-commercial research and educational use only. Not for reproduction, distribution or commercial use. This chapter was originally published in the book, The Enzymes, Vol .27, published by Elsevier, and the attached copy is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non-commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues who know you, and providing a copy to your institution’s administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution’s website or repository, are prohibited. For exceptions, permission may be sought for such use through Elsevier's permissions site at: http://www.elsevier.com/locate/permissionusematerial From: ESTELA JACINTO, AGC Kinases in mTOR Signaling, In Mike Hall and Fuyuhiko Tamanoi: The Enzymes, Vol. 27, Burlington: Academic Press, 2010, pp.101-128. ISBN: 978-0-12-381539-2, © Copyright 2010 Elsevier Inc, Academic Press. Author's personal copy 7 AGC Kinases in mTOR Signaling ESTELA JACINTO Department of Physiology and Biophysics UMDNJ-Robert Wood Johnson Medical School, Piscataway New Jersey, USA I. Abstract The mammalian target of rapamycin (mTOR), a protein kinase with homology to lipid kinases, orchestrates cellular responses to growth and stress signals. Various extracellular and intracellular inputs to mTOR are known. mTOR processes these inputs as part of two mTOR protein com- plexes, mTORC1 or mTORC2. Surprisingly, despite the many cellular functions that are linked to mTOR, there are very few direct mTOR substrates identified to date. -

Phosphatidylinositol-3-Kinase Related Kinases (Pikks) in Radiation-Induced Dna Damage
Mil. Med. Sci. Lett. (Voj. Zdrav. Listy) 2012, vol. 81(4), p. 177-187 ISSN 0372-7025 DOI: 10.31482/mmsl.2012.025 REVIEW ARTICLE PHOSPHATIDYLINOSITOL-3-KINASE RELATED KINASES (PIKKS) IN RADIATION-INDUCED DNA DAMAGE Ales Tichy 1, Kamila Durisova 1, Eva Novotna 1, Lenka Zarybnicka 1, Jirina Vavrova 1, Jaroslav Pejchal 2, Zuzana Sinkorova 1 1 Department of Radiobiology, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic 2 Centrum of Advanced Studies, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic. Received 5 th September 2012. Revised 27 th November 2012. Published 7 th December 2012. Summary This review describes a drug target for cancer therapy, family of phosphatidylinositol-3 kinase related kinases (PIKKs), and it gives a comprehensive review of recent information. Besides general information about phosphatidylinositol-3 kinase superfamily, it characterizes a DNA-damage response pathway since it is monitored by PIKKs. Key words: PIKKs; ATM; ATR; DNA-PK; Ionising radiation; DNA-repair ABBREVIATIONS therapy and radiation play a pivotal role. Since cancer is one of the leading causes of death worldwide, it is DSB - double stand breaks, reasonable to invest time and resources in the enligh - IR - ionising radiation, tening of mechanisms, which underlie radio-resis - p53 - TP53 tumour suppressors, tance. PI - phosphatidylinositol. The aim of this review is to describe the family INTRODUCTION of phosphatidyinositol 3-kinases (PI3K) and its func - tional subgroup - phosphatidylinositol-3-kinase rela - An efficient cancer treatment means to restore ted kinases (PIKKs) and their relation to repairing of controlled tissue growth via interfering with cell sig - radiation-induced DNA damage. -

Diverse Mechanisms Activate the PI 3-Kinase/Mtor Pathway In
Tran et al. BMC Cancer (2021) 21:136 https://doi.org/10.1186/s12885-021-07826-4 RESEARCH ARTICLE Open Access Diverse mechanisms activate the PI 3- kinase/mTOR pathway in melanomas: implications for the use of PI 3-kinase inhibitors to overcome resistance to inhibitors of BRAF and MEK Khanh B. Tran1,2,3, Sharada Kolekar1, Anower Jabed2, Patrick Jaynes2, Jen-Hsing Shih2, Qian Wang2, Jack U. Flanagan1,3, Gordon W. Rewcastle1,3, Bruce C. Baguley1,3 and Peter R. Shepherd1,2,3* Abstract Background: The PI 3-kinase (PI3K) pathway has been implicated as a target for melanoma therapy. Methods: Given the high degree of genetic heterogeneity in melanoma, we sought to understand the breadth of variation in PI3K signalling in the large NZM panel of early passage cell lines developed from metastatic melanomas. Results: We find the vast majority of lines show upregulation of this pathway, and this upregulation is achieved by a wide range of mechanisms. Expression of all class-IA PI3K isoforms was readily detected in these cell lines. A range of genetic changes in different components of the PI3K pathway was seen in different lines. Coding variants or amplification were identified in the PIK3CA gene, and amplification of the PK3CG gene was common. Deletions in the PIK3R1 and PIK3R2 regulatory subunits were also relatively common. Notably, no genetic variants were seen in the PIK3CD gene despite p110δ being expressed in many of the lines. Genetic variants were detected in a number of genes that encode phosphatases regulating the PI3K signalling, with reductions in copy number common in PTEN, INPP4B, INPP5J, PHLLP1 and PHLLP2 genes. -

Role of Cyclin-Dependent Kinase 1 in Translational Regulation in the M-Phase
cells Review Role of Cyclin-Dependent Kinase 1 in Translational Regulation in the M-Phase Jaroslav Kalous *, Denisa Jansová and Andrej Šušor Institute of Animal Physiology and Genetics, Academy of Sciences of the Czech Republic, Rumburska 89, 27721 Libechov, Czech Republic; [email protected] (D.J.); [email protected] (A.Š.) * Correspondence: [email protected] Received: 28 April 2020; Accepted: 24 June 2020; Published: 27 June 2020 Abstract: Cyclin dependent kinase 1 (CDK1) has been primarily identified as a key cell cycle regulator in both mitosis and meiosis. Recently, an extramitotic function of CDK1 emerged when evidence was found that CDK1 is involved in many cellular events that are essential for cell proliferation and survival. In this review we summarize the involvement of CDK1 in the initiation and elongation steps of protein synthesis in the cell. During its activation, CDK1 influences the initiation of protein synthesis, promotes the activity of specific translational initiation factors and affects the functioning of a subset of elongation factors. Our review provides insights into gene expression regulation during the transcriptionally silent M-phase and describes quantitative and qualitative translational changes based on the extramitotic role of the cell cycle master regulator CDK1 to optimize temporal synthesis of proteins to sustain the division-related processes: mitosis and cytokinesis. Keywords: CDK1; 4E-BP1; mTOR; mRNA; translation; M-phase 1. Introduction 1.1. Cyclin Dependent Kinase 1 (CDK1) Is a Subunit of the M Phase-Promoting Factor (MPF) CDK1, a serine/threonine kinase, is a catalytic subunit of the M phase-promoting factor (MPF) complex which is essential for cell cycle control during the G1-S and G2-M phase transitions of eukaryotic cells. -

Role of Ataxia-Telangiectasia Mutated Kinase in Cardiac Autophagy and Glucose Metabolism Under Ischemic Conditions Patsy Thrasher East Tennessee State University
East Tennessee State University Digital Commons @ East Tennessee State University Electronic Theses and Dissertations Student Works 8-2018 Role of Ataxia-Telangiectasia Mutated Kinase in Cardiac Autophagy and Glucose Metabolism Under Ischemic Conditions Patsy Thrasher East Tennessee State University Follow this and additional works at: https://dc.etsu.edu/etd Part of the Biology Commons, and the Physiology Commons Recommended Citation Thrasher, Patsy, "Role of Ataxia-Telangiectasia Mutated Kinase in Cardiac Autophagy and Glucose Metabolism Under Ischemic Conditions" (2018). Electronic Theses and Dissertations. Paper 3442. https://dc.etsu.edu/etd/3442 This Dissertation - Open Access is brought to you for free and open access by the Student Works at Digital Commons @ East Tennessee State University. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Digital Commons @ East Tennessee State University. For more information, please contact [email protected]. Role of Ataxia-Telangiectasia Mutated Kinase in Cardiac Autophagy and Glucose Metabolism Under Ischemic Conditions ____________________________________ A dissertation presented to the faculty of the Department of Biomedical Sciences East Tennessee State University In partial fulfillment of the requirements for the degree Doctor of Philosophy in Biomedical Sciences __________________________________ by Patsy R. Thrasher August 2018 _________________________________ Krishna Singh, Ph.D., Chair Mahipal Singh, Ph.D. Chuanfu Li, M.D. Tom Ecay, Ph.D. Yue Zou, Ph.D. Douglas Thewke, Ph.D. Keywords: ATM, myocardial infarction, autophagy, ischemia, glucose, metabolism ABSTRACT Role of Ataxia-Telangiectasia Mutated Kinase in Cardiac Autophagy and Glucose Metabolism Under Ischemic Conditions by Patsy R. Thrasher Ataxia-telangiectasia mutated kinase (ATM), a serine/threonine kinase primarily located in the nucleus, is typically activated in response to DNA damage. -

Characterization of the Small Molecule Kinase Inhibitor SU11248 (Sunitinib/ SUTENT in Vitro and in Vivo
TECHNISCHE UNIVERSITÄT MÜNCHEN Lehrstuhl für Genetik Characterization of the Small Molecule Kinase Inhibitor SU11248 (Sunitinib/ SUTENT in vitro and in vivo - Towards Response Prediction in Cancer Therapy with Kinase Inhibitors Michaela Bairlein Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzender: Univ. -Prof. Dr. K. Schneitz Prüfer der Dissertation: 1. Univ.-Prof. Dr. A. Gierl 2. Hon.-Prof. Dr. h.c. A. Ullrich (Eberhard-Karls-Universität Tübingen) 3. Univ.-Prof. A. Schnieke, Ph.D. Die Dissertation wurde am 07.01.2010 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 19.04.2010 angenommen. FOR MY PARENTS 1 Contents 2 Summary ................................................................................................................................................................... 5 3 Zusammenfassung .................................................................................................................................................... 6 4 Introduction .............................................................................................................................................................. 8 4.1 Cancer .............................................................................................................................................................. -

MAP2K4/MKK4 Expression in Pancreatic Cancer: Genetic Validation of Immunohistochemistry and Relationship to Disease Course
8516 Vol. 10, 8516–8520, December 15, 2004 Clinical Cancer Research MAP2K4/MKK4 Expression in Pancreatic Cancer: Genetic Validation of Immunohistochemistry and Relationship to Disease Course Wei Xin,1 Ki J. Yun,4 Francesca Ricci,2 ing patterns were also evaluated among unresectable pri- Marianna Zahurak,3 Wanglong Qiu,4 mary and metastatic cancer tissues from autopsy specimens, Gloria H. Su,4 Charles J. Yeo,5,6 indicating intact Mkk4 immunolabeling in 88.8% of the unresectable primary carcinomas as compared with 63.3% Ralph H. Hruban,4,5 Scott E. Kern,4,5 and 4,5 of distant metastases (P < 0.001). Our data indicate that the Christine A. Iacobuzio-Donahue loss of Mkk4 protein expression in pancreatic carcinomas 1 Department of Pathology, The University of Michigan Medical may be more frequent than suggested by the rates of genetic Center, Ann Arbor, Michigan; 2Department of Pathology, University La Sapienza, Rome, Italy; and the Departments of 3Biostatistics, inactivation alone and that MKK4 loss may contribute to 4Pathology, 5Oncology, and 6Surgery, The Johns Hopkins University disease progression. The correlation of MKK4 genetic status Hospital, Baltimore, Maryland with immunolabeling patterns validate this approach for the evaluation of MKK4 status in routine histologic sections and ABSTRACT may provide useful information regarding patient prognosis. MKK4 (MAP2K4/SEK1) is a member of the mitogen- activated protein kinase family, originally identified as a INTRODUCTION kinase involved in the stress-activated protein kinase path- The mitogen-activated protein (MAP) kinase cascades are way by directly phosphorylating c-Jun NH -terminal kinase. 2 multifunctional signaling pathways that are evolutionally well MKK4 genetic inactivation has been observed in a subset of conserved in all of the eukaryotic cells. -

Androgen Receptor
DAPAGLIFLOZIN ARGININE Sodium/glucoseSodium/glucose cotransporter cotransporter 2 1 TIPIFARNIB GANETESPIB ORVEPITANT REPARIXINNitric oxide synthase- inducible NORETHINDRONE DENUFOSOL TETRASODIUM Nitric-oxide synthase- brain NILOTINIB BREMELANOTIDE CANNABIDIOL GeranylgeranylHeat transferase shockHeat protein shock type proteinHSP I 90-alpha HSP 90-beta NAVITOCLAXSubstance-P receptor AFATINIB Protein farnesyltransferase Platelet activating factor receptor Estrogen receptorInterleukin-8 alphaICLAPRIMInterleukin-8 receptor receptor B A LASOFOXIFENE ORLISTAT Androgen Receptor Dihydrofolate reductaseCystic fibrosis transmembrane conductance regulator Melanocortin receptor 4 Nuclear factor ApoptosisNF-kappa-B regulator complex Bcl-XPyrimidinergic receptor P2Y6 NINTEDANIB Melanocortin receptorCannabinoid 3G-protein CB1 coupled receptor receptor 55 Purinergic receptor P2Y2 Vasopressin V2 receptor Estrogen receptor Apoptosis regulatorLUMACAFTOR Bcl-2 ERTEBERELTERIPARATIDE IVACAFTOR Melanocortin receptor 5 BETAMETHASONE Progesterone receptorEstrogen receptorBcl2-antagonist betaRUTIN of cell deathPyrimidinergic (BAD) receptor P2Y4 Cannabinoid CB2 receptor Parathyroid hormone receptor Vasopressin V1a receptorVASOPRESSIN CISPLATINNuclear receptor subfamily 1 group I member 2 COSYNTROPIN BICALUTAMIDE AZITHROMYCIN DOXYCYCLINE HYCLATE CytochromeIRINOTECAN P450 HYDROCHLORIDE2B6 PATUPILONE HYDRATE OXYTOCIN DEXTROTHYROXINEBETAMETHASONELORAZEPAM VALERATE Smoothened homolog LIXIVAPTAN TOLVAPTAN ARTEMISININ Glucagon-like peptide 1 receptor VANIPREVIR Regulator -

Mtor Signaling in Metabolism and Cancer
cells Editorial mTOR Signaling in Metabolism and Cancer Shile Huang 1,2 1 Department of Biochemistry and Molecular Biology, Louisiana State University Health Sciences Center, 1501 Kings Highway, Shreveport, LA 71130-3932, USA; [email protected]; Tel.: +1-318-675-7759 2 Feist-Weiller Cancer Center, Louisiana State University Health Sciences Center, 1501 Kings Highway, Shreveport, LA 71130-3932, USA Received: 10 October 2020; Accepted: 13 October 2020; Published: 13 October 2020 Abstract: The mechanistic/mammalian target of rapamycin (mTOR), a serine/threonine kinase, is a central regulator for human physiological activity. Deregulated mTOR signaling is implicated in a variety of disorders, such as cancer, obesity, diabetes, and neurodegenerative diseases. The papers published in this special issue summarize the current understanding of the mTOR pathway and its role in the regulation of tissue regeneration, regulatory T cell differentiation and function, and different types of cancer including hematologic malignancies, skin, prostate, breast, and head and neck cancer. The findings highlight that targeting the mTOR pathway is a promising strategy to fight against certain human diseases. Keywords: mTOR; PI3K; Akt; tissue regeneration; regulatory T cells; tumor; photodynamic therapy The mechanistic/mammalian target of rapamycin (mTOR), a serine/threonine kinase, integrates environmental cues such as hormones, growth factors, nutrients, oxygen, and energy, regulating cell growth, proliferation, survival, motility and differentiation as well as metabolism (reviewed in [1,2]). Evidence has demonstrated that deregulated mTOR signaling is implicated in a variety of disorders, such as cancer, obesity, diabetes, and neurodegenerative diseases (reviewed in [1,2]). Current knowledge indicates that mTOR functions at least as two distinct complexes (mTORC1 and mTORC2) in mammalian cells. -

Androgen Receptor
RALTITREXED Dihydrofolate reductase BORTEZOMIB IsocitrateCannabinoid dehydrogenase CB1EPIRUBICIN receptor HYDROCHLORIDE [NADP] cytoplasmic VINCRISTINE SULFATE Hypoxia-inducible factor 1 alpha DOXORUBICINAtaxin-2 HYDROCHLORIDENIFENAZONEFOLIC ACID PYRIMETHAMINECellular tumor antigen p53 Muscleblind-likeThyroidVINBURNINEVINBLASTINETRIFLURIDINE protein stimulating 1 DEQUALINIUM SULFATEhormone receptor CHLORIDE Menin/Histone-lysine N-methyltransferasePHENELZINE MLLLANATOSIDE SULFATE C MELATONINDAUNORUBICINBETAMETHASONEGlucagon-like HYDROCHLORIDEEndonuclease peptide 4 1 receptor NICLOSAMIDEDIGITOXINIRINOTECAN HYDROCHLORIDE HYDRATE BISACODYL METHOTREXATEPaired boxAZITHROMYCIN protein Pax-8 ATPase family AAA domain-containing proteinLIPOIC 5 ACID, ALPHA Nuclear receptorCLADRIBINEDIGOXIN ROR-gammaTRIAMTERENE CARMUSTINEEndoplasmic reticulum-associatedFLUOROURACIL amyloid beta-peptide-binding protein OXYPHENBUTAZONEORLISTAT IDARUBICIN HYDROCHLORIDE 6-phospho-1-fructokinaseHeat shockSIMVASTATIN protein beta-1 TOPOTECAN HYDROCHLORIDE AZACITIDINEBloom syndromeNITAZOXANIDE protein Huntingtin Human immunodeficiency virus typeTIPRANAVIR 1 protease VitaminCOLCHICINE D receptorVITAMIN E FLOXURIDINE TAR DNA-binding protein 43 BROMOCRIPTINE MESYLATEPACLITAXEL CARFILZOMIBAnthrax lethalFlap factorendonucleasePrelamin-A/C 1 CYTARABINE Vasopressin V2 receptor AMITRIPTYLINEMicrotubule-associated HYDROCHLORIDERetinoidTRIMETHOPRIM proteinMothers X receptor tau against alpha decapentaplegic homolog 3 Histone-lysine N-methyltransferase-PODOFILOX H3 lysine-9OXYQUINOLINE -
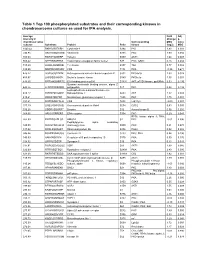
Table 1 Top 100 Phosphorylated Substrates and Their Corresponding Kinases in Chondrosarcoma Cultures As Used for IPA Analysis
Table 1 Top 100 phosphorylated substrates and their corresponding kinases in chondrosarcoma cultures as used for IPA analysis. Average Fold Adj intensity in Change p- chondrosarcoma Corresponding MSC value cultures Substrate Protein Psite kinase (log2) MSC 1043.42 RKKKVSSTKRH Cytohesin-1 S394 PKC 1.83 0.001 746.95 RKGYRSQRGHS Vitronectin S381 PKC 1.00 0.056 709.03 RARSTSLNERP Tuberin S939 AKT1 1.64 0.008 559.42 SPPRSSLRRSS Transcription elongation factor A-like1 S37 PKC; GSK3 0.18 0.684 515.29 LRRSLSRSMSQ Telethonin S157 Titin 0.77 0.082 510.00 MQPDNSSDSDY CD5 T434 PKA -0.35 0.671 476.27 GGRGGSRARNL Heterogeneous nuclear ribonucleoprotein K S302 PKCdelta 1.03 0.028 455.97 LKPGSSHRKTK Bruton's tyrosine kinase S180 PKCbeta 1.55 0.001 444.65 RRRMASMQRTG E1A binding protein p300 S1834 AKT; p70S6 kinase; pp90Rsk 0.53 0.195 Guanine nucleotide binding protein, alpha Z 440.26 HLRSESQRQRR polypeptide S27 PKC 0.88 0.199 6-phosphofructo-2-kinase/fructose-2,6- 424.12 RPRNYSVGSRP biphosphatase 2 S483 AKT 1.32 0.003 419.61 KKKIATRKPRF Metabotropic glutamate receptor 1 T695 PKC 1.75 0.001 391.21 DNSSDSDYDLH CD5 T453 Lck; Fyn -2.09 0.001 377.39 LRQLRSPRRAQ Ras associated protein Rab4 S204 CDC2 0.63 0.091 376.28 SSQRVSSYRRT Desmin S12 Aurora kinase B 0.56 0.255 369.05 ARIGGSRRERS EP4 receptor S354 PKC 0.29 0.543 RPS6 kinase alpha 3; PKA; 367.99 EPKRRSARLSA HMG14 S7 PKC -0.01 0.996 Peptidylglycine alpha amidating 349.08 SRKGYSRKGFD monooxygenase S930 PKC 0.21 0.678 347.92 RRRLSSLRAST Ribosomal protein S6 S236 PAK2 0.02 0.985 346.84 RSNPPSRKGSG Connexin -

Mtor Inhibitors Potentially Reduce TGF-Β2-Induced Fibrogenic
www.nature.com/scientificreports OPEN mTOR inhibitors potentially reduce TGF‑β2‑induced fbrogenic changes in trabecular meshwork cells Nozomi Igarashi, Megumi Honjo* & Makoto Aihara We examined the efects of mTOR inhibitors on the fbrotic response induced by transforming growth factor‑beta2 (TGF‑β2) in cultured human trabecular meshwork (hTM) cells. TGF‑β2‑induced expression of fbronectin, collagen type I, alpha 1 chain (COL1A1), and alpha‑smooth muscle actin (αSMA) in hTM cells was examined in the presence or absence of mTOR inhibitors using quantitative real‑time polymerase chain reaction, Western blotting, and immunohistochemistry. The migration rates of hTM cells were examined in the presence of TGF‑β2 with or without mTOR inhibitors. An in vitro study showed that the expression of fbronectin, COL1A1, and αSMA was upregulated by TGF‑β2 treatment of hTM cells; such upregulation was signifcantly suppressed by mTOR inhibitors. The inhibitors signifcantly reduced the migration rate of TGF‑β2‑stimulated hTM cells. mTOR inhibitors may usefully reduce the fbrotic response of hTM cells and we may have to explore if it is also efective in in vivo model. Glaucoma is known to be the second leading cause of blindness worldwide. Aberrant increases in intraocular pressure (IOP) characterize glaucoma, and such an increase in IOP can damage the optic nerve 1–3. IOP reduction is the only established efective therapy that suppresses visual impairment and blindness in both hypertensive and normotensive individuals. IOP elevation is reported to be principally the result of increased aqueous humor (AH) outfow resistance within the conventional outfow pathway 1,4,5. Damage to the collector channels is impli- cated as a cause of resistance within the distal pathway in patients with advanced glaucoma 6.