UC Riverside UC Riverside Previously Published Works
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Topic: Apomixis - Definition and Types
Course Teacher: Dr. Rupendra Kumar Jhade, JNKVV, College of Horticulture,Chhindwara(M.P.) B.Sc.(Hons) Horticulture- 1st Year, II Semester 2019-20 Course: Plant Propagation and Nursery Management Lecture No. 12 Topic: Apomixis - Definition and types Apomixis, derived from two Greek word “APO” (away from) and “MIXIS” (act of mixing or mingling). The first discovery of this phenomenon is credited to Leuwen hock as early as 1719 in Citrus seeds. In apomixes, seeds are formed but the embryos develop without fertilization. The genotype of the embryo and resulting plant will be the same as the seed parent. Definition: “The process of development of diploid embryos without fertilization.” Or “Apomixis is a form of asexual reproduction that occurs via seeds, in which embryos develop without fertilization.” Or “Apomixis is a type of reproduction in which sexual organs of related structures take part but seeds are formed without union of gametes.” In some species of plants, an embryo develops from the diploid cells of the seed and not as a result of fertilization between ovule and pollen. This type of reproduction is known as apomixis and the seedlings produced in this manner are known as apomicts. Apomictic seedlings are identical to mother plant and similar to plants raised by other vegetative means, because such plants have the same genetic make-up as that of the mother plant. Such seedlings are completely free from viruses. In apomictic species, sexual reproduction is either suppressed or absent. When sexual reproduction also occurs, the apomixes is termed as facultative apomixis. But when sexual reproduction is absent, it is referred to as obligate apomixis. -

Tropical Horticulture: Lecture 32 1
Tropical Horticulture: Lecture 32 Lecture 32 Citrus Citrus: Citrus spp., Rutaceae Citrus are subtropical, evergreen plants originating in southeast Asia and the Malay archipelago but the precise origins are obscure. There are about 1600 species in the subfamily Aurantioideae. The tribe Citreae has 13 genera, most of which are graft and cross compatible with the genus Citrus. There are some tropical species (pomelo). All Citrus combined are the most important fruit crop next to grape. 1 Tropical Horticulture: Lecture 32 The common features are a superior ovary on a raised disc, transparent (pellucid) dots on leaves, and the presence of aromatic oils in leaves and fruits. Citrus has increased in importance in the United States with the development of frozen concentrate which is much superior to canned citrus juice. Per-capita consumption in the US is extremely high. Citrus mitis (calamondin), a miniature orange, is widely grown as an ornamental house pot plant. History Citrus is first mentioned in Chinese literature in 2200 BCE. First citrus in Europe seems to have been the citron, a fruit which has religious significance in Jewish festivals. Mentioned in 310 BCE by Theophrastus. Lemons and limes and sour orange may have been mutations of the citron. The Romans grew sour orange and lemons in 50–100 CE; the first mention of sweet orange in Europe was made in 1400. Columbus brought citrus on his second voyage in 1493 and the first plantation started in Haiti. In 1565 the first citrus was brought to the US in Saint Augustine. 2 Tropical Horticulture: Lecture 32 Taxonomy Citrus classification based on morphology of mature fruit (e.g. -

Comparative Transcriptome Sequencing to Determine Genes Related to the Nucellar Embryony Mechanism in Citrus
Turkish Journal of Agriculture and Forestry Turk J Agric For (2019) 43: 58-68 http://journals.tubitak.gov.tr/agriculture/ © TÜBİTAK Research Article doi:10.3906/tar-1806-10 Comparative transcriptome sequencing to determine genes related to the nucellar embryony mechanism in citrus 1, 2 1 1 1 Özhan ŞİMŞEK *, Dicle DÖNMEZ , Sinan ETİ , Turgut YEŞİLOĞLU , Yıldız AKA KAÇAR 1 Department of Horticulture, Faculty of Agriculture, Çukurova University, Adana, Turkey 2 Biotechnology Research and Application Center, Çukurova University, Adana, Turkey Received: 04.06.2018 Accepted/Published Online: 14.10.2018 Final Version: 06.02.2019 Abstract: Several citrus varieties produce apomictic seeds by the nucellar embryony (NE) mechanism. Nucellar embryos are created from nucellus tissue and have an identical genetic constitution to the mother plant. Nucellar embryony is known to be an unusual feature of seed production in many citrus cultivars. The term “NE” refers to the development of identical embryos from the maternal tissue known as the nucellus surrounding the embryo sac. The authors aimed here to detect differentially expressed genes involved in the NE mechanism. Orlando tangelo (OT), producing apomictic seeds, and a clementine mandarin, Algerian tangerine ranch selection (AT), known as monoembryonic, were used for high-throughput transcriptome sequencing. First of all, histological analysis was used to determine the initial stage of the development of NE cells. Initial NE cells began to develop on the third day after anthesis. Based on the histological analysis, ovules of flower buds for OT were sampled at the balloon stage and 1, 3, and 5 days after anthesis; for AT only ovules of flower buds at the balloon stage and 3 days after anthesis were sampled for comparative transcriptome sequencing. -

Improvement of Subtropical Fruit Crops: Citrus
IMPROVEMENT OF SUBTROPICAL FRUIT CROPS: CITRUS HAMILTON P. ÏRAUB, Senior Iloriiciilturist T. RALPH ROBCNSON, Senior Physiolo- gist Division of Frnil and Vegetable Crops and Diseases, Bureau of Plant Tndusiry MORE than half of the 13 fruit crops known to have been cultivated longer than 4,000 years,according to the researches of DeCandolle (7)\ are tropical and subtropical fruits—mango, oliv^e, fig, date, banana, jujube, and pomegranate. The citrus fruits as a group, the lychee, and the persimmon have been cultivated for thousands of years in the Orient; the avocado and papaya were important food crops in the American Tropics and subtropics long before the discovery of the New World. Other types, such as the pineapple, granadilla, cherimoya, jaboticaba, etc., are of more recent introduction, and some of these have not received the attention of the plant breeder to any appreciable extent. Through the centuries preceding recorded history and up to recent times, progress in the improvement of most subtropical fruits was accomplished by the trial-error method, which is crude and usually expensive if measured by modern standards. With the general accept- ance of the Mendelian principles of heredity—unit characters, domi- nance, and segregation—early in the twentieth century a starting point was provided for the development of a truly modern science of genetics. In this article it is the purpose to consider how subtropical citrus fruit crops have been improved, are now being improved, or are likel3^ to be improved by scientific breeding. Each of the more important crops will be considered more or less in detail. -

Citrus Rootstocks: Their Characters and Reactions
CITRUS ROOTSTOCKS: THEIR CHARACTERS AND REACTIONS (an unpublished manuscript) ca. 1986 By W. P. BITTERS (1915 – 2006) Editor, digital version: Marty Nemeth, Reference Librarian, UC Riverside Science Library, retired Subject matter experts, digital version: Dr. Tracy Kahn, Curator, UC Citrus Variety Collection Dr. Robert Krueger, Curator, USDA-ARS National Clonal Germplasm Repository for Citrus & Dates Toni Siebert, Assistant Curator, UC Citrus Variety Collection ca. 1955 ca. 1970 IN MEMORIUM Willard P. Bitters Professor of Horticulture, Emeritus Riverside 1915-2006 Born in Eau Claire, Wisconsin, in June, 1915, Dr. Willard “Bill” Bitters earned his bachelor’s degree in biology from St. Norbert College and his master’s degree and Ph.D. from the University of Wisconsin. After earning his doctorate, he first worked as the superintendent of the Valley Research Farm of the University of Arizona in Yuma, and joined the Citrus Experiment Station, in Riverside in 1946 as a Horticulturist. In 1961, Dr. Bitters became a Professor in the newly established University of California-Riverside. His initial assignment was to work on horticultural aspects of tristeza, a serious vector-transmitted virus disease which threatened to destroy California citrus orchards. Tristeza was already in California and spreading in 1946. At that time most citrus trees in California were grafted on a rootstock that was known to be susceptible to tristeza. Dr. Bill Bitters was responsible for screening of over 500 cultivars to determine which rootstock-scion combinations were resistant to this disease and yet possessed suitable horticultural characteristics. Of the 500 screened, most were susceptible, but several successful ones were selected and released to the industry. -

Genomic Analyses of Primitive, Wild and Cultivated Citrus Provide Insights Into Asexual Reproduction
ARTICLES OPEN Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction Xia Wang1,7, Yuantao Xu1,7, Siqi Zhang1,7, Li Cao2,7, Yue Huang1, Junfeng Cheng3, Guizhi Wu1, Shilin Tian4, Chunli Chen5, Yan Liu3, Huiwen Yu1, Xiaoming Yang1, Hong Lan1, Nan Wang1, Lun Wang1, Jidi Xu1, Xiaolin Jiang1, Zongzhou Xie1, Meilian Tan1, Robert M Larkin1, Ling-Ling Chen3, Bin-Guang Ma3, Yijun Ruan5,6, Xiuxin Deng1 & Qiang Xu1 The emergence of apomixis—the transition from sexual to asexual reproduction—is a prominent feature of modern citrus. Here we de novo sequenced and comprehensively studied the genomes of four representative citrus species. Additionally, we sequenced 100 accessions of primitive, wild and cultivated citrus. Comparative population analysis suggested that genomic regions harboring energy- and reproduction-associated genes are probably under selection in cultivated citrus. We also narrowed the genetic locus responsible for citrus polyembryony, a form of apomixis, to an 80-kb region containing 11 candidate genes. One of these, CitRWP, is expressed at higher levels in ovules of polyembryonic cultivars. We found a miniature inverted-repeat transposable element insertion in the promoter region of CitRWP that cosegregated with polyembryony. This study provides new insights into citrus apomixis and constitutes a promising resource for the mining of agriculturally important genes. Asexual reproduction is a remarkable feature of perennial fruit crops important trait for breeding purposes. On the one hand, polyembryony that facilitates the faithful propagation of commercially valuable is widely employed in citrus nurseries and propagation programs to individuals by avoiding the uncertainty associated with the sexual generate large numbers of uniform rootstocks from seeds. -

A Pattern of Unique Embryogenesis Occurring Via Apomixis in Carya Cathayensis
BIOLOGIA PLANTARUM 56 (4): 620-627, 2012 DOI: 10.1007/s10535-012-0256-2 A pattern of unique embryogenesis occurring via apomixis in Carya cathayensis B. ZHANG1,2a, Z.J. WANG1a, S.H. JIN1a, G.H. XIA1, Y.J. HUANG1 and J.Q. HUANG1* School of Forestry and Biotechnology, Zhejiang A & F University, Lin’an 311300, P.R. China1 Bureau of Agricultural Economics of Haiyan, Haiyan 314300, P.R. China2 Abstract Apomixis represents an alteration of classical sexual plant reproduction to produce seeds that have essentially clonal embryos. In this report, hickory (Carya cathayensis Sarg.), which is an important oil tree, is identified as a new apomictic species. The ovary has a chamber containing one ovule that is unitegmic and orthotropous. Embryological investigations indicated that the developmental pattern of embryo sac formation is typical polygonum-type. Zygote embryos were not found during numerous histological investigations, and the embryo originated from nucellar cells. Nucellar embryo initials were found both at the micropylar and chalazal ends of the embryo sac, but the mature embryo developed only at the nucellar beak region. The mass of the nucellar embryo initial at the nucellar beak region developed into a nucellar embryo or split into two nucellar proembryos. The later development of the nucellar embryo was similar to the zygotic embryo and progressed from globular embryo to heart-shape embryo and to cotyledon embryo. Additional key words: adventitious embryony, female gametophyte, hickory, nucellar embryo. Introduction The term apomixis was first introduced by Winkler 3) two gametophytic types diplospory and apospory (1908) to refer to the “substitution of sexual reproduction (Peggy 2006, Koltunow 1993). -
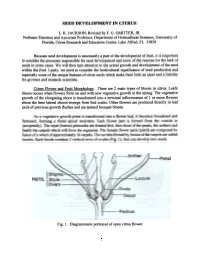
Seed Development in Citrus
SEED DEVELOPMENT IN CITRUS L. K. JACKSON; Revisedby F. G. GMITTER, JR. ProfessorEmeritus and AssociateProfessor, Department of Horticultural Sciences,University of Florida, Citrus Researchand EducationCenter, Lake Alfred, FL 33850 Becauseseed development is necessarilya part of the developmentof fruit. it is important to considerthe processesresponsible for seeddevelopment and someof the reasonsfor the lack of seedsin somecases. We will then turn attentionto the actual growth and developmentof the seed within the fruit. Lastly, we needto considerthe horticultural significanceof seedproduction and especiallysome of the uniquefeatures of citrus seedswhich makethem both an assetand a liability for growersand researchscientists. Citrus Flower and Fruit MoQ1holo~. There are 2 main types of bloom in citrus. Leafy bloom occurswhen flowers form on and with new vegetativegrowth in the spring. The vegetative growth of the elongating shoot is transformedinto a tenninal inflorescenceof 1 or more flowers about the time lateral shootsemerge from bud scales.Other flowers are produceddirectly in leaf axils of previous growth flushesand are termedbouquet bloom. As a vegetative growth point is transfonned into a flower bud, it becomes broadened and flattened, forming a floral apical meristem. Each flower part is fonned from the outside in (acropetally). The sepal (button) primordia are fonned first, then those of the petals, the anthers and finally the carpels which will form the segments. The female flower parts (pistil) are composed by fusion of a whorl of approximately 10 carpels. The cavities fonned by fusion of the carpels are called locules. Each locule contains 2 vertical rows of ovules (Fig. 1), that can develop into seeds. Fig. 1. Diagrammaticportrayal of opencitrus flower. -

Citrus Genetic Resources in California
Citrus Genetic Resources in California Analysis and Recommendations for Long-Term Conservation Report of the Citrus Genetic Resources Assessment Task Force T.L. Kahn, R.R. Krueger, D.J. Gumpf, M.L. Roose, M.L. Arpaia, T.A. Batkin, J.A. Bash, O.J. Bier, M.T. Clegg, S.T. Cockerham, C.W. Coggins Jr., D. Durling, G. Elliott, P.A. Mauk, P.E. McGuire, C. Orman, C.O. Qualset, P.A. Roberts, R.K. Soost, J. Turco, S.G. Van Gundy, and B. Zuckerman Report No. 22 June 2001 Published by Genetic Resources Conservation Program Division of Agriculture and Natural Resources UNIVERSITY OF CALIFORNIA i This report is one of a series published by the University of California Genetic Resources Conservation Program (technical editor: P.E. McGuire) as part of the public information function of the Program. The Program sponsors projects in the collection, inventory, maintenance, preservation, and utilization of genetic resources important for the State of California as well as research and education in conservation biology. Further information about the Program may be obtained from: Genetic Resources Conservation Program University of California One Shields Avenue Davis, CA 95616 USA (530) 754-8501 FAX (530) 754-8505 e-mail: [email protected] Website: http://www.grcp.ucdavis.edu/ Additional copies of this report may be ordered from this address. Citation: Kahn TL, RR Krueger, DJ Gumpf, ML Roose, ML Arpaia, TA Batkin, JA Bash, OJ Bier, MT Clegg, ST Cockerham, CW Coggins Jr, D Durling, G Elliott, PA Mauk, PE McGuire, C Orman, CO Qualset, PA Roberts, RK Soost, J Turco, SG Van Gundy, and B Zuckerman. -

Fertilization and Development of Embryo on Satsuma Orange (Citrus Unshiu MARC.) and Natsudaidai (C
Fertilization and development of embryo on Satsuma orange (Citrus unshiu MARC.) and Natsudaidai (C. natsudaidai HAYATA) HSU-JENYANG Collegeof Agriculture,University of OsakaPrefecture , Sakai,Osaka Summary The mechanism of the Citrus pollen-tube growth 3. The fertilized eggs and nucellar cells began into style and ovary, fertilization and development to divide 40 to 50 days after pollination both in of embryo were studied. The results obtained were selfed Natsudaidai and crossed Satsuma orange with summarized as follows Natsudaidai. 1. The pollen tubes of Natsudaidai grew faster 4. In Natsudaidai, embryo development was very than Satsuma orange in pistil of Natsudaidai or slow and reached the globular stage 58 days, heart- Satsuma orange. shape embryo stage 72 days, and the complete 2. The fertilization took place about 8 days in embryo stage 120 days after self-pollination. selfed Natsudaidai, 12 days in the crossing of 5. The nucellar cells of one to three layers near Satsuma orange with Natsudaidai, and 14 days in the micropylar of embryo sac and the region of self-pollination of Satsuma orange after pollination, the micropylar end to one-third of embryo sac have respectively. the ability to form nucellar embryos. Introduction The polyembryony has been found extensively in Citrus, Poncirus and Fortunella plants. This phenomenon was first recognized in orange seed by LEEVWENHOEKin the beginning of the eighteenth century. According to the works of OSAWA(19) ToxopEUs (21) FROST5, FURUSATO(7) and MAHESHWARIand SACHAR~I5~,the extra embryo originate from the nucellar tissue in ovules. It has been considered that pollination is positively necessary for the occurrence of the nucellar embryos (IKEDA(11~,NAGAI and TANIKAWA{17~,FROST~4~, WONG(23)), however, WRIHGT~24~,reported that nucellar embryos occur even without pollination. -

From Forest to Field: Perennial Fruit Crop Domestication
American Journal of Botany 98(9): 1389–1414. 2011. SPECIAL PAPER F ROM FOREST TO FIELD: PERENNIAL FRUIT CROP DOMESTICATION 1 Allison J. Miller 2,4 and Briana L. Gross3 2 Department of Biology, Saint Louis University, 3507 Laclede Avenue, Saint Louis, Missouri 63103 USA; and 3 USDA-ARS, National Center for Genetic Resource Preservation, 1111 S. Mason Street, Fort Collins, Colorado 80521 USA • Premise of the study: Archaeological and genetic analyses of seed-propagated annual crops have greatly advanced our under- standing of plant domestication and evolution. Comparatively little is known about perennial plant domestication, a relevant topic for understanding how genes and genomes evolve in long-lived species, and how perennials respond to selection pres- sures operating on a relatively short time scale. Here, we focus on long-lived perennial crops (mainly trees and other woody plants) grown for their fruits. • Key results: We reviewed (1) the basic biology of long-lived perennials, setting the stage for perennial domestication by con- sidering how these species evolve in nature; (2) the suite of morphological features associated with perennial fruit crops under- going domestication; (3) the origins and evolution of domesticated perennials grown for their fruits; and (4) the genetic basis of domestication in perennial fruit crops. • Conclusions: Long-lived perennials have lengthy juvenile phases, extensive outcrossing, widespread hybridization, and limited population structure. Under domestication, these features, combined with clonal propagation, multiple origins, and ongoing crop – wild gene fl ow, contribute to mild domestication bottlenecks in perennial fruit crops. Morphological changes under do- mestication have many parallels to annual crops, but with key differences for mating system evolution and mode of reproduc- tion. -

PROBLEMS in FRUIT BREEDING - POLYPLOIDY and HETEROZYGOSITY Polyploidy an Organism Having More Than Two Sets of Homologous Chromosomes Is Known As a Polyploid
PROBLEMS IN FRUIT BREEDING - POLYPLOIDY AND HETEROZYGOSITY Polyploidy An organism having more than two sets of homologous chromosomes is known as a polyploid. Polyploidy is of general occurrence in plants while it is rare amongst animals. If the somatic chromosome sets in a diploid be represented by AA BB CC then the genome, i.e., the number in the genomes will be A B C. If this is represented by ‘n’ then the simple polyploid series would be: 2n – diploid 3n – triploid 4n – tetraploid 5n – pentaploid 6n – hexaploid 7n – heptaploid 8n – octaploid 9n – Nonaploid 10n – decaploid and so on Polyploidy is pervasive in plants and some estimates suggest that 30-80% of living plant species are polyploids, and many lineage show evidence of ancient polyploidy (paleopolyploidy) in their genomes. Polyploid plants can arise spontaneously in nature by several mechanisms, including meiotic or mitotic failures, and fusion of unreduced (2n) gametes. Both autopolyploids (e.g. Potato) and allopolyploids (e.g. canola, wheat and cotton) can be found among both wild and domesticated plant species. Most polyploids display heterosis relative to their parental species. The mechanisms leading to novel variation in newly formed allopolyploids may include gene dosage effects (resulting from more numerous copies of genome content), the reunion of divergent gene regulatory hierarchies, chromosomal rearrangements, and epigenetic remodeling, all of which affect gene content and or expression levels. Many of these rapid changes contribute to reproductive isolation and speciation. Behaviour of polyploid crops Polyploid plants tend to be larger and better at thriving in early succession habitats such as farm fields.