Usability of Nuclear Single-Copy Genes Compared with Plastid DNA on Different Phylogenetic Levels of and Within the Order Poales
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Published Vestigations Together Study Existing Accept Arrangements
Notes on the Nomenclature of some grasses II by Dr. J.Th. Henrard (Rijksherbarium, Leiden) (Issued September 10th, 1941). In a former article new combinations and critical observa- 1) many all the world. New in- tions were published on various grasses over vestigations in critical genera together with the study of the existing literature made it necessary to accept various other arrangements in this important family. The old system of Bentham, once the basis for a total is and modified and review, now more more many tribes are and limited. The have purified more exactly most recent system we at the moment, is Hubbard’s treatment of this family in the work of Hutchinson: The families of flowering plants. Vol. II. Monocotyle- dons. The grasses are divided there into 26 tribes. We have here the great advantage that aberrant which are into genera, not easy to place one of the formerly accepted tribes, are given as representatives of distinct new tribes. The curious tropical genus Streptochaeta f.i. con- stitutes the tribe of the Streptochaeteae. It is quite acceptable that tribes consist of but may one genus, especially when such a genus is a totally deviating one and cannot be inserted into one of the already existing ones. Such tribes are f.i. the Nardeae with the only northern genus Nardus, and the Mediterranean tribe of the Lygeeae with the only genus Lygeum, one of the Esparto grasses. It is therefore wonder no that Hubbard creates a new tribe, the Anomochloeae, for one of the most curious tropical grasses of the world. -
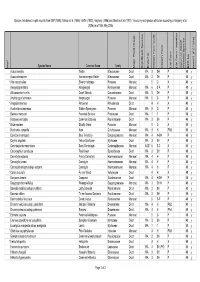
BFS048 Site Species List
Species lists based on plot records from DEP (1996), Gibson et al. (1994), Griffin (1993), Keighery (1996) and Weston et al. (1992). Taxonomy and species attributes according to Keighery et al. (2006) as of 16th May 2005. Species Name Common Name Family Major Plant Group Significant Species Endemic Growth Form Code Growth Form Life Form Life Form - aquatics Common SSCP Wetland Species BFS No kens01 (FCT23a) Wd? Acacia sessilis Wattle Mimosaceae Dicot WA 3 SH P 48 y Acacia stenoptera Narrow-winged Wattle Mimosaceae Dicot WA 3 SH P 48 y * Aira caryophyllea Silvery Hairgrass Poaceae Monocot 5 G A 48 y Alexgeorgea nitens Alexgeorgea Restionaceae Monocot WA 6 S-R P 48 y Allocasuarina humilis Dwarf Sheoak Casuarinaceae Dicot WA 3 SH P 48 y Amphipogon turbinatus Amphipogon Poaceae Monocot WA 5 G P 48 y * Anagallis arvensis Pimpernel Primulaceae Dicot 4 H A 48 y Austrostipa compressa Golden Speargrass Poaceae Monocot WA 5 G P 48 y Banksia menziesii Firewood Banksia Proteaceae Dicot WA 1 T P 48 y Bossiaea eriocarpa Common Bossiaea Papilionaceae Dicot WA 3 SH P 48 y * Briza maxima Blowfly Grass Poaceae Monocot 5 G A 48 y Burchardia congesta Kara Colchicaceae Monocot WA 4 H PAB 48 y Calectasia narragara Blue Tinsel Lily Dasypogonaceae Monocot WA 4 H-SH P 48 y Calytrix angulata Yellow Starflower Myrtaceae Dicot WA 3 SH P 48 y Centrolepis drummondiana Sand Centrolepis Centrolepidaceae Monocot AUST 6 S-C A 48 y Conostephium pendulum Pearlflower Epacridaceae Dicot WA 3 SH P 48 y Conostylis aculeata Prickly Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis juncea Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis setigera subsp. -

INDIAN RICEGRASS Erosion Control/Reclamation: One of Indian Ricegrass' Greatest Values Is for Stabilizing Sites Susceptible to Achnatherum Hymenoides Wind Erosion
Plant Guide INDIAN RICEGRASS Erosion control/reclamation: One of Indian ricegrass' greatest values is for stabilizing sites susceptible to Achnatherum hymenoides wind erosion. It is well adapted to stabilization of (Roemer & J.A. Schultes) disturbed sandy soils in mixes with other species. It is naturally an early invader onto disturbed sandy Barkworth sites (after and in concert with needle and thread Plant Symbol = ACHY grass). It is also one of the first to establish on cut and fill slopes. It does not compete well with Contributed By: USDA NRCS Idaho State Office aggressive introduced grasses during the establishment period, but is very compatible with slower developing natives, such as Snake River wheatgrass (Elymus wawawaiensis), bluebunch wheatgrass (Pseudoroegneria spicata), thickspike wheatgrass (Elymus lanceolata ssp. lanceolata), streambank wheatgrass (Elymus lanceolata ssp. psammophila), western wheatgrass (Pascopyrum smithii), and needlegrass species (Stipa spp. and Ptilagrostis spp.). Drought tolerance combined with fibrous root system and fair to good seedling vigor, make Indian ricegrass desirable for reclamation in areas receiving 8 to 14 inches annual precipitation. Wildlife: Forage value is mentioned in the grazing/rangeland/hayland section above. Due to the abundance of plump, nutritious seed produced by Indian ricegrass, it is considered an excellent food source for birds, such as morning doves, pheasants, and songbirds. Rodents collect the seed for winter food supplies. It is considered good cover habitat for small -

Special Issue3.7 MB
Volume Eleven Conservation Science 2016 Western Australia Review and synthesis of knowledge of insular ecology, with emphasis on the islands of Western Australia IAN ABBOTT and ALLAN WILLS i TABLE OF CONTENTS Page ABSTRACT 1 INTRODUCTION 2 METHODS 17 Data sources 17 Personal knowledge 17 Assumptions 17 Nomenclatural conventions 17 PRELIMINARY 18 Concepts and definitions 18 Island nomenclature 18 Scope 20 INSULAR FEATURES AND THE ISLAND SYNDROME 20 Physical description 20 Biological description 23 Reduced species richness 23 Occurrence of endemic species or subspecies 23 Occurrence of unique ecosystems 27 Species characteristic of WA islands 27 Hyperabundance 30 Habitat changes 31 Behavioural changes 32 Morphological changes 33 Changes in niches 35 Genetic changes 35 CONCEPTUAL FRAMEWORK 36 Degree of exposure to wave action and salt spray 36 Normal exposure 36 Extreme exposure and tidal surge 40 Substrate 41 Topographic variation 42 Maximum elevation 43 Climate 44 Number and extent of vegetation and other types of habitat present 45 Degree of isolation from the nearest source area 49 History: Time since separation (or formation) 52 Planar area 54 Presence of breeding seals, seabirds, and turtles 59 Presence of Indigenous people 60 Activities of Europeans 63 Sampling completeness and comparability 81 Ecological interactions 83 Coups de foudres 94 LINKAGES BETWEEN THE 15 FACTORS 94 ii THE TRANSITION FROM MAINLAND TO ISLAND: KNOWNS; KNOWN UNKNOWNS; AND UNKNOWN UNKNOWNS 96 SPECIES TURNOVER 99 Landbird species 100 Seabird species 108 Waterbird -

KLAUS AMMANN, BIBLIOGRAPHY PUBLIC PAPERS TIL 20199423 [email protected]
KLAUS AMMANN, BIBLIOGRAPHY PUBLIC PAPERS TIL 20199423 [email protected] Adenle Ademola and Ammann Klaus (2015) Role of Modern Biotechnology in Sustainable Development; Addressing Social-Political Dispute of GMOs that Influences Decision-Making in Developing countries UNITED NATIONS, Economic and Social Affairs New York, USA 3 pp https://sustainabledevelopment.un.org/content/documents/1758GSDR%202015%20Advance%20Unedi ted%20Version.pdf AND printed: http://www.ask-force.org/web/Sustainability/Adenle-Ammann-Role- Modern-Biotech-Sustainable-2015.pdf AND manuscript with full text links: http://www.ask- force.org/web/Sustainability/Adenle-Ammann-Role-Modern-Biotech-Sustainable-fullltext-2015.pdf Adenle Ademola, Morris Jane E. and Murphy Denis J. (2017) Genetically Modified Organisms in Developing Countries: Risk Analysis and Governance 1st Edition, Kindle Edition Cambridge University Press; 1 edition (May 31, 2017 Adenle Ademola, Morris Jane E. and Murphy Denis J. Book Genetically Modified Organisms in Developing Countries Kindl edition, Cambridge, UK 300 pp ISBN: 1107151910 AND ASIN: B0722LJ5M7/ISBN: 1107151910 AND ASIN: B0722LJ5M7 https://www.amazon.com/Genetically-Modified-Organisms-Developing-Countries- ebook/dp/B0722LJ5M7/ref=sr_1_1_twi_kin_2?s=books&ie=UTF8&qid=1499199684&sr=1- 1&keywords=Adenle+Ademola Bibl. Ka, contents http://www.ask-force.org/web/Developing/Adenle- Morris-Murphy-CONTENTS-Genetically-modified-organisms-in-developing-countries-risk-analysis-and- governance-2017.pdf Altmann Michael and Ammann Klaus (1991) Die -

Poaceae: Pooideae) Based on Plastid and Nuclear DNA Sequences
d i v e r s i t y , p h y l o g e n y , a n d e v o l u t i o n i n t h e monocotyledons e d i t e d b y s e b e r g , p e t e r s e n , b a r f o d & d a v i s a a r h u s u n i v e r s i t y p r e s s , d e n m a r k , 2 0 1 0 Phylogenetics of Stipeae (Poaceae: Pooideae) Based on Plastid and Nuclear DNA Sequences Konstantin Romaschenko,1 Paul M. Peterson,2 Robert J. Soreng,2 Núria Garcia-Jacas,3 and Alfonso Susanna3 1M. G. Kholodny Institute of Botany, Tereshchenkovska 2, 01601 Kiev, Ukraine 2Smithsonian Institution, Department of Botany MRC-166, National Museum of Natural History, P.O. Box 37012, Washington, District of Columbia 20013-7012 USA. 3Laboratory of Molecular Systematics, Botanic Institute of Barcelona (CSIC-ICUB), Pg. del Migdia, s.n., E08038 Barcelona, Spain Author for correspondence ([email protected]) Abstract—The Stipeae tribe is a group of 400−600 grass species of worldwide distribution that are currently placed in 21 genera. The ‘needlegrasses’ are char- acterized by having single-flowered spikelets and stout, terminally-awned lem- mas. We conducted a molecular phylogenetic study of the Stipeae (including all genera except Anemanthele) using a total of 94 species (nine species were used as outgroups) based on five plastid DNA regions (trnK-5’matK, matK, trnHGUG-psbA, trnL5’-trnF, and ndhF) and a single nuclear DNA region (ITS). -

Vicariance, Climate Change, Anatomy and Phylogeny of Restionaceae
Botanical Journal of the Linnean Society (2000), 134: 159–177. With 12 figures doi:10.1006/bojl.2000.0368, available online at http://www.idealibrary.com on Under the microscope: plant anatomy and systematics. Edited by P. J. Rudall and P. Gasson Vicariance, climate change, anatomy and phylogeny of Restionaceae H. P. LINDER FLS Bolus Herbarium, University of Cape Town, Rondebosch 7701, South Africa Cutler suggested almost 30 years ago that there was convergent evolution between African and Australian Restionaceae in the distinctive culm anatomical features of Restionaceae. This was based on his interpretation of the homologies of the anatomical features, and these are here tested against a ‘supertree’ phylogeny, based on three separate phylogenies. The first is based on morphology and includes all genera; the other two are based on molecular sequences from the chloroplast genome; one covers the African genera, and the other the Australian genera. This analysis corroborates Cutler’s interpretation of convergent evolution between African and Australian Restionaceae. However, it indicates that for the Australian genera, the evolutionary pathway of the culm anatomy is much more complex than originally thought. In the most likely scenario, the ancestral Restionaceae have protective cells derived from the chlorenchyma. These persist in African Restionaceae, but are soon lost in Australian Restionaceae. Pillar cells and sclerenchyma ribs evolve early in the diversification of Australian Restionaceae, but are secondarily lost numerous times. In some of the reduction cases, the result is a very simple culm anatomy, which Cutler had interpreted as a primitively simple culm type, while in other cases it appears as if the functions of the ribs and pillars may have been taken over by a new structure, protective cells developed from epidermal, rather than chlorenchyma, cells. -

5 Priority Flora (Comesperma Rhadinocarpum, Desmocladus Elongatus, Hemiandra Sp
PO Box 437 Kalamunda WA 6926 +61 08 9257 1625 [email protected] (ACN 063 507 175, ABN 39 063 507 175) INTRODUCTION The following brief overview was prepared to assist in outlining some of the issues for planning rehabilitation needs on the proposed Silica sands operations by VRX Silica. As a summary of key points it raises some issues that need discussion with the operational team at VRX Silica. VEGETATION DIRECT TRANSFER (VDT) TRIAL A rehabilitation technique that was first implemented at Iluka Eneabba in 2012 by the Iluka site personnel. Since this time the Mattiske Consulting team have been assessing the progress of the plants from this method. As such it supplements the extensive rehabilitation practices on the Iluka operations which relies on the more classical approach of site preparation and seeding. Iluka differs from other rehabilitation areas on mining operations in that the native vegetation was also mulched and applied to the rehabilitation areas. The rehabilitation technique of VDT, or community translocation, is the practice of salvaging and replacing intact sods of vegetation with the underlying soil intact (Ross et al, 2000). The method trialled on the VDT 16 trial Vegetation Direct Transfer transects in 2019. The use of this technique by Iluka in the former Jennings mining area aims to improve the sustainability of translocating largely recalcitrant sedge and rush species, which tend to be less well represented in rehabilitation via other techniques, due to their low or complete lack of seed production (Norman and Koch 2007), but which often dominate local heath communities. The use of deep profile direct return of the topsoil and overburden in one pass may provide a large scale method of translocating rhizomatous and tuberous species in rehabilitated areas (Norman and Koch 2007). -

Hans Walter Lack & Thomas Raus Hildemar Scholz
Fl. Medit. 22: 233-244 doi: 10.7320/FlMedit22.233 Version of Record published online on 28 December 2012 Hans Walter Lack & Thomas Raus Hildemar Scholz (1928 – 2012) Hildemar Scholz, aged 84, passed away on 5 June 2012 as a consequence of a bad fall he had suffered at home a few weeks earlier. He was a world authority on grasses, the adventive flora of Europe, and rust fungi from central Europe. From 1964 until his retire- ment in 1993 he was a member of staff of the Botanic Garden and Botanical Museum Berlin-Dahlem, rising from scientific collaborator to one of the directors at this institution. Hildemar was a born plant-collector active all over Europe, in North and West Africa with a special interest in grasses and the rust fungi parasitizing them. Born on 27 May 1928 in Berlin into a family of protestant clergymen Hildemar Scholz remained both in language and manners very much a Berliner, in the true and best sense of the word, and never ever considered leaving his home town, even during the many diffi- cult years this city encountered in the last century. Having belatedly finished his school training because of the Second World War in 1947, he tried to get enrolled at Berlin University, subsequently renamed Humboldt University, in the Soviet sector of the city, but was rejected – probably because his father was a clergyman. Hildemar had more success at the newly founded Free University in the American sector of Berlin, where he got admit- ted in 1949 and chose biology as his subject. -

Poaceae) Author(S): Raúl Gonzalo , Carlos Aedo , and Miguel Ángel García Source: Systematic Botany, 38(2):344-378
Taxonomic Revision of the Eurasian Stipa Subsections Stipa and Tirsae (Poaceae) Author(s): Raúl Gonzalo , Carlos Aedo , and Miguel Ángel García Source: Systematic Botany, 38(2):344-378. 2013. Published By: The American Society of Plant Taxonomists URL: http://www.bioone.org/doi/full/10.1600/036364413X666615 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Systematic Botany (2013), 38(2): pp. 344–378 © Copyright 2013 by the American Society of Plant Taxonomists DOI 10.1600/036364413X666615 Taxonomic Revision of the Eurasian Stipa Subsections Stipa and Tirsae (Poaceae) Rau´ l Gonzalo,1,2 Carlos Aedo,1 and Miguel A´ ngel Garcı´a1 1Real Jardı´n Bota´nico, CSIC, Dpto. de Biodiversidad y Conservacio´n. Plaza de Murillo 2, 28014 Madrid, Spain. 2Author for correspondence ([email protected]) Communicating Editor: Lucia G. Lohmann Abstract—A comprehensive taxonomic revision of Stipa subsects. -

(Poaceae: Panicoideae) in Thailand
Systematics of Arundinelleae and Andropogoneae, subtribes Chionachninae, Dimeriinae and Germainiinae (Poaceae: Panicoideae) in Thailand Thesis submitted to the University of Dublin, Trinity College for the Degree of Doctor of Philosophy (Ph.D.) by Atchara Teerawatananon 2009 Research conducted under the supervision of Dr. Trevor R. Hodkinson School of Natural Sciences Department of Botany Trinity College University of Dublin, Ireland I Declaration I hereby declare that the contents of this thesis are entirely my own work (except where otherwise stated) and that it has not been previously submitted as an exercise for a degree to this or any other university. I agree that library of the University of Dublin, Trinity College may lend or copy this thesis subject to the source being acknowledged. _______________________ Atchara Teerawatananon II Abstract This thesis has provided a comprehensive taxonomic account of tribe Arundinelleae, and subtribes Chionachninae, Dimeriinae and Germainiinae of the tribe Andropogoneae in Thailand. Complete floristic treatments of these taxa have been completed for the Flora of Thailand project. Keys to genera and species, species descriptions, synonyms, typifications, illustrations, distribution maps and lists of specimens examined, are also presented. Fourteen species and three genera of tribe Arundinelleae, three species and two genera of subtribe Chionachninae, seven species of subtribe Dimeriinae, and twelve species and two genera of Germainiinae, were recorded in Thailand, of which Garnotia ciliata and Jansenella griffithiana were recorded for the first time for Thailand. Three endemic grasses, Arundinella kerrii, A. kokutensis and Dimeria kerrii were described as new species to science. Phylogenetic relationships among major subfamilies in Poaceae and among major tribes within Panicoideae were evaluated using parsimony analysis of plastid DNA regions, trnL-F and atpB- rbcL, and a nuclear ribosomal DNA region, ITS. -

Data Standards Version 2.8 July 5
Euro+Med Data Standards Version 2.8. July 5th, 2002 EURO+MED PLANTBASE PREPARATION OF THE INITIAL CHECKLIST: DATA STANDARDS VERSION 2.8 JULY 5TH, 2002 This document replaces Version 2.7, dated May 16th, 2002 Compiled for the Euro+Med PlantBase Editorial Committee by: Euro+Med PlantBase Secretariat, Centre for Plant Diversity and Systematics, School of Plant Sciences, The University of Reading, Whiteknights, Reading RG6 6AS United Kingdom Tel: +44 (0)118 9318160 Fax: +44 (0)118 975 3676 E-mail: [email protected] 1 Euro+Med Data Standards Version 2.8. July 5th, 2002 Modifications made in Version 2.0 (24/11/00) 1. Section 2.4 as been corrected to note that geography should be added for hybrids as well as species and subspecies. 2. Section 3 (Standard Floras) has been modified to reflect the presently accepted list. This may be subject to further modification as the project proceeds. 3. Section 4 (Family Blocks) – genera have been listed where this clarifies the circumscription of blocks. 4. Section 5 (Accented Characters) – now included in the document with examples. 5. Section 6 (Geographical Standard) – Macedonia (Mc) is now listed as Former Yugoslav Republic of Macedonia. Modification made in Version 2.1 (10/01/01) Page 26: Liliaceae in Block 21 has been corrected to Lilaeaceae. Modifications made in Version 2.2 (4/5/01) Geographical Standards. Changes made as discussed at Palermo General meeting (Executive Committee): Treatment of Belgium and Luxembourg as separate areas Shetland not Zetland Moldova not Moldavia Czech Republic