Poaceae: Pooideae) Based on Plastid and Nuclear DNA Sequences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Vascular Plants of Massachusetts
The Vascular Plants of Massachusetts: The Vascular Plants of Massachusetts: A County Checklist • First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Somers Bruce Sorrie and Paul Connolly, Bryan Cullina, Melissa Dow Revision • First A County Checklist Plants of Massachusetts: Vascular The A County Checklist First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Massachusetts Natural Heritage & Endangered Species Program Massachusetts Division of Fisheries and Wildlife Natural Heritage & Endangered Species Program The Natural Heritage & Endangered Species Program (NHESP), part of the Massachusetts Division of Fisheries and Wildlife, is one of the programs forming the Natural Heritage network. NHESP is responsible for the conservation and protection of hundreds of species that are not hunted, fished, trapped, or commercially harvested in the state. The Program's highest priority is protecting the 176 species of vertebrate and invertebrate animals and 259 species of native plants that are officially listed as Endangered, Threatened or of Special Concern in Massachusetts. Endangered species conservation in Massachusetts depends on you! A major source of funding for the protection of rare and endangered species comes from voluntary donations on state income tax forms. Contributions go to the Natural Heritage & Endangered Species Fund, which provides a portion of the operating budget for the Natural Heritage & Endangered Species Program. NHESP protects rare species through biological inventory, -

View Plant List Here
12th annual Theodore Payne Native Plant Garden Tour Plant List GARDEN 5 in pasadena provided by homeowner Botanical Name Common Name Abutilon palmeri Indian Mallow Achillea millefolium ‘Calistoga’ Calistoga Yarrow Achillea millefolium ‘Island Pink’ Island Pink Yarrow Achnatherum--see Stipa Aesculus californica California Buckeye Arctostaphylos glauca Big Berry Manzanita Arctostaphylos silvicola ‘Ghostly’ Ghostly Manzanita Artemisia californica California Sagebrush Artemisia ludoviciana ‘Silver King’ Silver King Wormwood Artemisia tridentata Great Basin Sagebrush Asclepias eriocarpa Kotolo or Indian Milkweed Astragalus pychnostachyus var. lanosissimus Ventura Marsh Milkvetch Bahiopsis (Viguiera) laciniata San Diego Sunflower Berberis (Mahonia) nervosa Cascades Oregon Grape Bergerocactus emoryi Cunyado, Golden Spined Cereus Brahea armata Mexican Blue Palm Calystegia macrostegia ‘Anacapa Pink’ Anacapa Pink Island Morning Glory Carpenteria californica Bush Anemone Dendromecon harfordii Channel Island Bush Poppy Diplacus (Mimulus) aurantiacus var. puniceus ‘Torrey Torrey Pines Red Bush Monkeyflower Pines’ Dudleya anomala no common name Dudleya pulverulenta Chalk Dudleya Encelia californica California Bush Sunflower Encelia farinosa Brittlebush, Incienso Epilobium (Zauschneria) ‘Silver Select’ Silver Select California Fuchsia Eriogonum fasciculatum ‘Dana Point’ Dana Point California Buckwheat Fouquieria splendens Ocotillo Frangula (Rhamnus) californica ‘Eve Case’ Eve Case Coffeeberry Gutierrezia californica California Matchweed Hazardia detonsa -

2020 Majura Ainslie Plant List.Xlsx
Plant Species List for Mount Majura and Mount Ainslie, Canberra Base data from Ingwerson, F; O. Evans & B. Griffiths. (1974). Vegetation of the Ainslie-Majura Reserve . Conservation Series No. 2. AGPS Canberra. Re-organised, revised and updated by Michael Doherty, CSIRO Ecosystem Sciences and Waltraud Pix, Friends of Mt. Majura With advice from Isobel Crawford, Australian Botanical Surveys Current version of 01.10.2020 Names: Census of Plants of the Australian Capital Territory, Version 4.1, 2019 Enquiries:Version 3.0 [email protected] (8th June 2012) subsp. = subspecies Form ? = questionable status or identity f = herb, forb sp. aff. = having close affinities with i.e. similar but not quite the sameo = herb, orchid syn. = synonymous with i.e. most recent previous name, or alternativeg = nameherb, grass sens. lat. = in the broad sense of the species concept gl = herb, grass- or sedge-like var. = variety s = shrub (including creeper and climber) sp. = species i.e. identity yet to be finalised st = shrub / small tree spp. = species in the plural i.e. more than one species t = tree MM Mount Majura. Notionally north of “Blue Metal” Road; MA Mount Ainslie. Notionally south of “Blue Metal” Road (VVV) Species occurrence checking; currently focused on Mt. Majura rather than Mt. Ainslie. No ticks next to name = species reported but not yet confirmed for Mt Majura and Mt Ainslie. Status is locally native except for: PE = Planted Exotic PN = Planted Non-local Native WE = Weed Exotic WN = Weed Non-local Native ‘Planted’ status refers to individuals which are planted but not spreading ‘Weed’ status refers to species reproducing in the wild Scientific name Common name MM MA Status Form Family Isolepis sp . -
BFS048 Site Species List
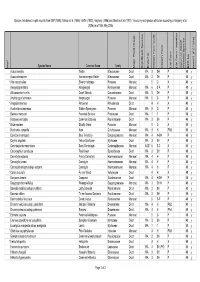
Species lists based on plot records from DEP (1996), Gibson et al. (1994), Griffin (1993), Keighery (1996) and Weston et al. (1992). Taxonomy and species attributes according to Keighery et al. (2006) as of 16th May 2005. Species Name Common Name Family Major Plant Group Significant Species Endemic Growth Form Code Growth Form Life Form Life Form - aquatics Common SSCP Wetland Species BFS No kens01 (FCT23a) Wd? Acacia sessilis Wattle Mimosaceae Dicot WA 3 SH P 48 y Acacia stenoptera Narrow-winged Wattle Mimosaceae Dicot WA 3 SH P 48 y * Aira caryophyllea Silvery Hairgrass Poaceae Monocot 5 G A 48 y Alexgeorgea nitens Alexgeorgea Restionaceae Monocot WA 6 S-R P 48 y Allocasuarina humilis Dwarf Sheoak Casuarinaceae Dicot WA 3 SH P 48 y Amphipogon turbinatus Amphipogon Poaceae Monocot WA 5 G P 48 y * Anagallis arvensis Pimpernel Primulaceae Dicot 4 H A 48 y Austrostipa compressa Golden Speargrass Poaceae Monocot WA 5 G P 48 y Banksia menziesii Firewood Banksia Proteaceae Dicot WA 1 T P 48 y Bossiaea eriocarpa Common Bossiaea Papilionaceae Dicot WA 3 SH P 48 y * Briza maxima Blowfly Grass Poaceae Monocot 5 G A 48 y Burchardia congesta Kara Colchicaceae Monocot WA 4 H PAB 48 y Calectasia narragara Blue Tinsel Lily Dasypogonaceae Monocot WA 4 H-SH P 48 y Calytrix angulata Yellow Starflower Myrtaceae Dicot WA 3 SH P 48 y Centrolepis drummondiana Sand Centrolepis Centrolepidaceae Monocot AUST 6 S-C A 48 y Conostephium pendulum Pearlflower Epacridaceae Dicot WA 3 SH P 48 y Conostylis aculeata Prickly Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis juncea Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis setigera subsp. -

Missouriensis
Missouriensis Journal of the Missouri Native Plant Society Volume 34 2017 effectively published online 30 September 2017 Missouriensis, Volume 34 (2017) Journal of the Missouri Native Plant Society EDITOR Douglas Ladd Missouri Botanical Garden P.O. Box 299 St. Louis, MO 63110 email: [email protected] MISSOURI NATIVE PLANT SOCIETY https://monativeplants.org PRESIDENT VICE-PRESIDENT John Oliver Dana Thomas 4861 Gatesbury Drive 1530 E. Farm Road 96 Saint Louis, MO 63128 Springfield, MO 65803 314.487.5924 317.430.6566 email: [email protected] email: [email protected] SECRETARY TREASURER Malissa Briggler Bob Siemer 102975 County Rd. 371 74 Conway Cove Drive New Bloomfield, MO 65043 Chesterfield, MO 63017 573.301.0082 636.537.2466 email: [email protected] email: [email protected] IMMEDIATE PAST PRESIDENT WEBMASTER Paul McKenzie Brian Edmond 2311 Grandview Circle 8878 N Farm Road 75 Columbia, MO 65203 Walnut Grove, MO 65770 573.445.3019 417.742.9438 email: [email protected] email: [email protected] BOARD MEMBERS Steve Buback, St. Joseph (2015-2018); email: [email protected] Ron Colatskie, Festus (2016-2019); email: [email protected] Rick Grey, St. Louis (2015-2018); email: [email protected] Bruce Schuette, Troy (2016-2019); email: [email protected] Mike Skinner, Republic (2016-2019); email: [email protected] Justin Thomas, Springfield (2014-2017); email: [email protected] i FROM THE EDITOR Welcome to the first online edition of Missouriensis. The format has been redesigned to facilitate access and on-screen readability, and articles are freely available online as open source, archival pdfs. -

BSBI News Index 121-130 ABC 8Pt FINAL
BSBI News INDEX to Nos 121 – 130 September 2012 to September 2015 Compiled by GWYNN ELLIS ISSN 2397-8813 1 GUIDE TO THE INDEX ABBREVIATIONS AEM Annual Exhibition Meeting Illus. Illustration AGM Annual General Meeting Infl. Inflorescence ASM Annual Summer Meeting Lvs Leaves cf. confer (compare) photo © photo copyright holder congrats congratulations Rev. Review CS Colour Section Rpt Report del. delineavit (drawn) s.l. sensu lato (broad sense) Descr. Description s.s. sensu stricto (narrow sense) Diag. Diagram v.c. vice-county Exbn Exhibition v.cc. vice-counties Exbt Exhibit (♀) female parent Fld Mtg Rpt Field Meeting Report (♂) male parent Fls Flowers ACKNOWLEDGEMENTS: The compiler wishes to thank David Pearman for much helpful advice and for scrutinising the final text. However, responsibility for checking the index and its final form rests solely with the compiler. BOOKS et al. are italicised as are Periodicals and scientific names COLOUR PAGES: In the index all colour page numbers are distinguished by being underlined with the cover pages enclosed in square brackets [ ]. The front cover and inside front cover are numbered [i] and [ii] respectively while the inside back and back cover pages are numbered according to the number of pages, thus with an issue of 76 pages the inside back cover is [77] and the back cover [78]. Colour Section plates are numbered CS1, CS2, CS3, CS4. Photographers are now indexed by name with the qualification (photo ©) COMPILATION: Using the original text on computer, the entries for each issue were generated by deleting all unwanted text. After checking, the entries were then sorted into alphabetical order, condensed, and finally output as pdf files for the Printer. -

Floerkea Proserpinacoides Willdenow False Mermaid-Weed
New England Plant Conservation Program Floerkea proserpinacoides Willdenow False Mermaid-weed Conservation and Research Plan for New England Prepared by: William H. Moorhead III Consulting Botanist Litchfield, Connecticut and Elizabeth J. Farnsworth Senior Research Ecologist New England Wild Flower Society Framingham, Massachusetts For: New England Wild Flower Society 180 Hemenway Road Framingham, MA 01701 508/877-7630 e-mail: [email protected] • website: www.newfs.org Approved, Regional Advisory Council, December 2003 1 SUMMARY Floerkea proserpinacoides Willdenow, false mermaid-weed, is an herbaceous annual and the only member of the Limnanthaceae in New England. The species has a disjunct but widespread range throughout North America, with eastern and western segregates separated by the Great Plains. In the east, it ranges from Nova Scotia south to Louisiana and west to Minnesota and Missouri. In the west, it ranges from British Columbia to California, east to Utah and Colorado. Although regarded as Globally Secure (G5), national ranks of N? in Canada and the United States indicate some uncertainly about its true conservation status in North America. It is listed as rare (S1 or S2) in 20% of the states and provinces in which it occurs. Floerkea is known from only 11 sites total in New England: three historic sites in Vermont (where it is ranked SH), one historic population in Massachusetts (where it is ranked SX), and four extant and three historic localities in Connecticut (where it is ranked S1, Endangered). The Flora Conservanda: New England ranks it as a Division 2 (Regionally Rare) taxon. Floerkea inhabits open or forested floodplains, riverside seeps, and limestone cliffs in New England, and more generally moist alluvial soils, mesic forests, springy woods, and streamside meadows throughout its range. -

Vascular Plants and a Brief History of the Kiowa and Rita Blanca National Grasslands
United States Department of Agriculture Vascular Plants and a Brief Forest Service Rocky Mountain History of the Kiowa and Rita Research Station General Technical Report Blanca National Grasslands RMRS-GTR-233 December 2009 Donald L. Hazlett, Michael H. Schiebout, and Paulette L. Ford Hazlett, Donald L.; Schiebout, Michael H.; and Ford, Paulette L. 2009. Vascular plants and a brief history of the Kiowa and Rita Blanca National Grasslands. Gen. Tech. Rep. RMRS- GTR-233. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 44 p. Abstract Administered by the USDA Forest Service, the Kiowa and Rita Blanca National Grasslands occupy 230,000 acres of public land extending from northeastern New Mexico into the panhandles of Oklahoma and Texas. A mosaic of topographic features including canyons, plateaus, rolling grasslands and outcrops supports a diverse flora. Eight hundred twenty six (826) species of vascular plant species representing 81 plant families are known to occur on or near these public lands. This report includes a history of the area; ethnobotanical information; an introductory overview of the area including its climate, geology, vegetation, habitats, fauna, and ecological history; and a plant survey and information about the rare, poisonous, and exotic species from the area. A vascular plant checklist of 816 vascular plant taxa in the appendix includes scientific and common names, habitat types, and general distribution data for each species. This list is based on extensive plant collections and available herbarium collections. Authors Donald L. Hazlett is an ethnobotanist, Director of New World Plants and People consulting, and a research associate at the Denver Botanic Gardens, Denver, CO. -

Developing Species-Habitat Relationships: 2016 Project Report
Field Keys to Groups and Alliances in the National Vegetation Classification: Northern Basin & Range / Columbia Plateau Ecoregions NatureServe Conservation Science Division P r i n c i p a l Investigator Patrick J. C o m e r , Chief Ecologist [email protected] 703.797.4802 November 2017 Photos (clockwise from top left; all used under Creative Commons license CC BY 2.0.): Big sage shrubland, Humboldt-Toiyabe National Forest, Nevada. USDA Photo by Susan Elliot. http://flic.kr/p/ax64DY Jeffrey pine woodland, photo by David Prasad. https://www.flickr.com/photos/33671002@N00 Northwest Great Plains Mixedgrass Prairie, Dakota Prairie National Grasslands, North Dakota. Western juniper woodland, BLM Black Hills Recreation Area, Oregon. Acknowledgements This work was completed with funding provided by the Bureau of Land Management through the BLM’s Fish, Wildlife and Plant Conservation Resource Management Program under Cooperative Agreement L13AC00286 between NatureServe and the BLM. Suggested citation: Schulz, K., G. Kittel, M. Reid and P. Comer. 2017. Field Keys to Divisions, Macrogroups, Groups and Alliances in the National Vegetation Classification: Northern Basin & Range / Columbia Plateau Ecoregions. Report prepared for the Bureau of Land Management by NatureServe, Arlington VA. 14p + 58p of Keys + Appendices. See appendix document: Descriptions_NVC_Groups_Alliances_ NorthernBasinRange_Nov_2017.pdf 2 | P a g e Contents Introduction and Background ...................................................................................................................... -

TESIS Caracterización Ambiental De Los Ecosistemas, Zonas De Vida Y
UNIVERSIDAD NACIONAL DANIEL ALCIDES CARRIÓN FACULTAD DE INGENIERÍA ESCUELA DE FORMACIÓN PROFESIONAL DE INGENIERÍA AMBIENTAL TESIS Caracterización Ambiental De Los Ecosistemas, Zonas De Vida Y Vegetación Natural De La Provincia De Pasco Para optar el título profesional de: Ingeniero Ambiental Autor: Bach. Yelitza Orializ VILLEGAS ALANIA Asesor: Ing. Anderson MARCELO MANRIQUE Cerro de Pasco – Perú – 2019 UNIVERSIDAD NACIONAL DANIEL ALCIDES CARRIÓN FACULTAD DE INGENIERÍA ESCUELA DE FORMACIÓN PROFESIONAL DE INGENIERÍA AMBIENTAL TESIS Caracterización Ambiental De Los Ecosistemas, Zonas De Vida Y Vegetación Natural De La Provincia De Pasco Sustentada y aprobada ante los miembros del jurado: Mg. Julio Antonio ASTO LIÑAN Mg. Luis Alberto PACHECO PEÑA PRESIDENTE MIEMBRO Mg. David Johnny CUYUBAMBA ZEVALLOS MIEMBRO 2 DEDICATORIA A mi abuelita, madre y hermanos por el apoyo constante, el empeño y la confianza que me dieron durante el desarrollo y culminación de mis estudios A mi alma mater por brindarme la oportunidad de llegar a ser una profesional 3 RECONOCIMIENTO A Dios por permitirnos tener una buena experiencia dentro de la universidad, por permitir convertimos en profesionales en lo que tanto nos apasiona A mi abuelita, madre y hermanos quienes me brindaron su valiosa y desinteresada orientación y guía en la elaboración del presente trabajo de investigación A cada docente que fue parte de este proceso integral de formación profesional, ya que también fueron un apoyo para poder llegar a esta etapa de nuestras vidas Y a todas las personas que en una u otra forma me apoyaron en la realización de este trabajo 4 RESUMEN La superficie de la Provincia de Pasco abarca dos Eco regiones de las 21 identificadas para nuestro país: La puna de los Andes Centrales y la jalca, las cuales involucran grandes paisajes naturales, con características de formaciones vegetales de matorrales y también de praderas abiertas de gramíneas en los “pajonales” altos andinos. -

Vegetation Classification and Distribution Mapping Report: Hubbell Trading Post National Historic Site
National Park Service U.S. Department of the Interior Natural Resource Program Center Vegetation Classification and Distribution Mapping Report Hubbell Trading Post National Historic Site Natural Resource Technical Report NPS/SCPN/NRTR—2010/301 ON THE COVER Top: Hubbell Trading Post National Historic Site as seen from Hubbell Hill; photo by Courtney White, www.awestthatworks.com. Bottom left: Hubbell Trading Post National Historic Site; photo by Stephen Monroe. Bottom right: Hubbell Wash, photo by Stephen Monroe. Vegetation Classification and Distribution Mapping Report Hubbell Trading Post National Historic Site Natural Resource Technical Report NPS/SCPN/NRTR—2010/301 Authors David Salas Corey Bolen Bureau of Reclamation Remote Sensing and GIS Group Mail Code 86-68211 Denver Federal Center Building 67 Denver, Colorado 80225 Project Manager Anne Cully National Park Service, Southern Colorado Plateau Network P.O. Box 5765 Northern Arizona University Flagstaff, Arizona 86011 Editing and Design Jean Palumbo National Park Service, Southern Colorado Plateau Network P.O. Box 5765 Northern Arizona University Flagstaff, Arizona 86011 March 2010 U.S. Department of the Interior National Park Service Natural Resource Program Center Fort Collins, Colorado The National Park Service, Natural Resource Program Center publishes a range of reports that address natural resource topics of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituen cies, and the public. The Natural Resource Technical Report Series is used to disseminate results of scientific studies in the physical, biological, and social sciences for both the advancement of science and the achievement of the National Park Service mission. -

TEMPORAL and SPATIAL VARIATION in ALKALOID LEVELS in Achnatherum Robustum, a NATIVE GRASS INFECTED with the ENDOPHYTE Neotyphodium
Journal of Chemical Ecology, Vol. 32, No. 2, February 2006 ( #2006) DOI: 10.1007/s10886-005-9003-x TEMPORAL AND SPATIAL VARIATION IN ALKALOID LEVELS IN Achnatherum robustum, A NATIVE GRASS INFECTED WITH THE ENDOPHYTE Neotyphodium STANLEY H. FAETH,1,* DALE R. GARDNER,2 CINNAMON J. HAYES,1 ANDREA JANI,1 SALLY K. WITTLINGER,1 and THOMAS A. JONES3 1School of Life Sciences, Arizona State University, PO Box 874501 Tempe, AZ 85287-4501, USA 2USDA<ARS Poisonous Plant Research Laboratory, 1150 East 1400 North, Logan, UT 84341, USA 3USDA<ARS Forage and Range Research Laboratory, Utah State University, Logan, UT 84322, USA (Received August 5, 2005; revised October 4, 2005; accepted October 12, 2005) Published Online March 23, 2006 Abstract—The native North American perennial grass Achnatherum robus- tum (Vasey) Barkworth [= Stipa robusta (Vasey) Scribn.] or sleepygrass is toxic and narcotic to livestock. The causative agents are alkaloidal mycotoxins produced from infections by a systemic and asexual Neo- typhodium endophyte. Recent studies suggest that toxicity is limited across the range of sleepygrass in the Southwest USA. We sampled 17 populations of sleepygrass with varying distance from one focal population known for its high toxicity levels near Cloudcroft, NM, USA. For some, we sampled individual plants twice within the same growing season and over successive years (2001–2004). We also determined infection levels in each population. In general, all populations were highly infected, but infection levels were more variable near the focal population. Only infected plants within populations near the Cloudcroft area produced alkaloids. The ergot alkaloid, ergonovine, comprised the bulk of the alkaloids, with lesser amounts of lysergic and isolysergic acid amides and ergonovinine alkaloids.