LADON, a Natural Antisense Transcript of NODAL, Promotes Metastasis in Melanoma by Repressing NDRG1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
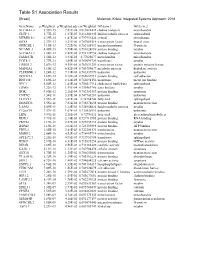
Table S1 Association Results
GeneName p.Weighted p.Weighted.adj cor.Weighted GO.term.1 GO.term.2 SLC44A1.2 3.55E-15 7.81E-08 0.819822422 choline transport mitochondria GLTP.1 5.77E-15 1.27E-07 0.816302445 lipid metabolic process sphingolipid MTMR10.1 6.39E-14 1.41E-06 0.797951424 cytosol phosphatase SOX8 2.37E-13 5.21E-06 0.787085514 transcription factor neural crest GPRC5B.1 4.19E-13 9.22E-06 0.782154937 integral membrane G-protein NCAM1.3 4.45E-13 9.79E-06 0.781624896 protein binding myelin SLC44A1.1 1.28E-12 2.82E-05 0.772132954 choline transport mitochondria FAM107B 1.56E-12 3.43E-05 0.77025677 mitochondria mitochondria UGT8.1 1.77E-12 3.89E-05 0.769099729 transferase myelin ERBB3.2 2.07E-12 4.55E-05 0.767631259 transcription factor protein tyrosine kinase MAN2A1 3.10E-12 6.82E-05 0.763759677 metabolic process hydrolase activity PLEKHH1.1 3.24E-12 7.13E-05 0.763337879 unknown unknown DOCK5.3 3.69E-12 8.12E-05 0.762087913 protein binding cell adhesion RNF130 3.69E-12 8.12E-05 0.762094156 membrane metal ion binding NPC1 6.50E-12 1.43E-04 0.756517114 cholesterol trafficking sphingolipid ERMN 7.22E-12 1.59E-04 0.755469786 actin binding myelin BOK 9.80E-12 2.16E-04 0.752383357 protein binding apoptosis CNTN2 1.54E-11 3.39E-04 0.747743281 unknown unknown ELOVL1 1.55E-11 3.41E-04 0.74764744 fatty acid sphingolipid DBNDD2 3.55E-11 7.81E-04 0.738878658 protein binding neuron projection LASS2 5.09E-11 1.12E-03 0.734954024 lipid metabolic process myelin C12orf34 7.57E-11 1.67E-03 0.730528911 unknown unknown LIPA 9.59E-11 2.11E-03 0.72786111 fatty acid glycerolipid metabolic -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype
Supplementary Information High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype Håkon Reikvam 1,2,*, Elise Aasebø 1, Annette K. Brenner 2, Sushma Bartaula-Brevik 1, Ida Sofie Grønningsæter 2, Rakel Brendsdal Forthun 2, Randi Hovland 3,4 and Øystein Bruserud 1,2 1 Department of Clinical Science, University of Bergen, 5020, Bergen, Norway 2 Department of Medicine, Haukeland University Hospital, 5021, Bergen, Norway 3 Department of Medical Genetics, Haukeland University Hospital, 5021, Bergen, Norway 4 Institute of Biomedicine, University of Bergen, 5020, Bergen, Norway * Correspondence: [email protected]; Tel.: +55-97-50-00 J. Clin. Med. 2019, 8, x 2 of 36 Figure S1. Mutational studies in a cohort of 71 AML patients. The figure shows the number of patients with the various mutations (upper), the number of mutations in for each patient (middle) and the number of main classes with mutation(s) in each patient (lower). 2 J. Clin. Med. 2019, 8, x; doi: www.mdpi.com/journal/jcm J. Clin. Med. 2019, 8, x 3 of 36 Figure S2. The immunophenotype of primary human AML cells derived from 62 unselected patients. The expression of the eight differentiation markers CD13, CD14, CD15, CD33, CD34, CD45, CD117 and HLA-DR was investigated for 62 of the 71 patients included in our present study. We performed an unsupervised hierarchical cluster analysis and identified four patient main clusters/patient subsets. The mutational profile for each f the 62 patients is also given (middle), no individual mutation of main class of mutations showed any significant association with any of the for differentiation marker clusters (middle). -
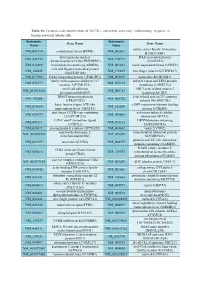
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

In Cancer Cells by Heat Shock Stimulation
583-593.qxd 19/7/2010 11:13 Ì ™ÂÏ›‰·583 INTERNATIONAL JOURNAL OF ONCOLOGY 37: 583-593, 2010 583 Induction of HITS, a newly identified family with sequence similarity 107 protein (FAM107B), in cancer cells by heat shock stimulation HIDEO NAKAJIMA1, YASUHITO ISHIGAKI2, QI-SHENG XIA1, TAKAYUKI IKEDA3, YOSHINO YOSHITAKE3, HIDETO YONEKURA3, TAKAYUKI NOJIMA4, TAKUJI TANAKA4, HISANORI UMEHARA5, NAOHISA TOMOSUGI2, TAKANOBU TAKATA2, TAKEO SHIMASAKI1, NAOKI NAKAYA1, ITARU SATO1, KAZUYUKI KAWAKAMI6, KEITA KOIZUMI7, TOSHINARI MINAMOTO6 and YOSHIHARU MOTOO1 1Department of Medical Oncology, 2Medical Research Institute and Departments of 3Biochemistry, 4Pathology and 5Hematology and Immunology, Kanazawa Medical University, 1-1 Daigaku, Uchinada, Kahoku, Ishikawa 920-0293; 6Division of Translational and Clinical Oncology, Cancer Research Institute and 7Department of Biophysical Genetics, Graduate School of Medical Science, Kanazawa University, 13-1 Takara-machi, Kanazawa, Ishikawa 920-0934, Japan Received March 31, 2010; Accepted May 14, 2010 DOI: 10.3892/ijo_00000707 Abstract. The Family with sequence similarity 107 (FAM107) Introduction possesses an N-terminal domain of unknown function (DUF1151) that is highly conserved beyond species. In human, The Family with sequence similarity 107 (FAM107) possesses FAM107A termed TU3A/DRR1 has been reported as a can- an N-terminal domain of unknown function (DUF1151) that didate tumor suppressor gene which expression is down- is conserved beyond species including mammalian, Xenopus, regulated in several types of cancer, however no studies have fish and Drosophila, and shows no homology match to investigated the other family protein, FAM107B. In the other functional conserved domains (http://www.ncbi.nlm. present study, we designated FAM107B as heat shock- nih.gov/structure/cdd/cdd.shtml). -

Heparanase Overexpression Induces Glucagon Resistance and Protects
Page 1 of 85 Diabetes Heparanase overexpression induces glucagon resistance and protects animals from chemically-induced diabetes Dahai Zhang1, Fulong Wang1, Nathaniel Lal1, Amy Pei-Ling Chiu1, Andrea Wan1, Jocelyn Jia1, Denise Bierende1, Stephane Flibotte1, Sunita Sinha1, Ali Asadi2, Xiaoke Hu2, Farnaz Taghizadeh2, Thomas Pulinilkunnil3, Corey Nislow1, Israel Vlodavsky4, James D. Johnson2, Timothy J. Kieffer2, Bahira Hussein1 and Brian Rodrigues1 1Faculty of Pharmaceutical Sciences, UBC, 2405 Wesbrook Mall, Vancouver, BC, Canada V6T 1Z3; 2Department of Cellular & Physiological Sciences, Life Sciences Institute, UBC, 2350 Health Sciences Mall, Vancouver, BC, Canada V6T 1Z3; 3Department of Biochemistry and Molecular Biology, Faculty of Medicine, Dalhousie University, 100 Tucker Park Road, Saint John, NB, Canada E2L 4L5; 4Cancer and Vascular Biology Research Center, Rappaport Faculty of Medicine, Technion, Haifa, Israel 31096 Running Title: Heparanase overexpression and the pancreatic islet Corresponding author: Dr. Brian Rodrigues Faculty of Pharmaceutical Sciences University of British Columbia, 2405 Wesbrook Mall, Vancouver, B.C., Canada V6T 1Z3 TEL: (604) 822-4758; FAX: (604) 822-3035 E-mail: [email protected] Key Words: Heparanase, heparan sulfate proteoglycan, glucose homeostasis, glucagon resistance, pancreatic islet, STZ Word Count: 4761 Total Number of DiabetesFigures: Publish 6 Ahead of Print, published online October 7, 2016 Diabetes Page 2 of 85 Abstract Heparanase, a protein with enzymatic and non-enzymatic properties, contributes towards disease progression and prevention. In the current study, a fortuitous observation in transgenic mice globally overexpressing heparanase (hep-tg) was the discovery of improved glucose homeostasis. We examined the mechanisms that contribute towards this improved glucose metabolism. Heparanase overexpression was associated with enhanced GSIS and hyperglucagonemia, in addition to changes in islet composition and structure. -

Amy Elizabeth Defnet Contact Information: [email protected] Degree and Date to Be Conferred: Ph.D
Targeting the Activator Protein-1 Complex to Inhibit Airway Smooth Muscle Cell Hyperproliferation in Asthma Item Type dissertation Authors Defnet, Amy Elizabeth Publication Date 2021 Abstract Hyperproliferation of airway smooth muscle (ASM) cells leads to increased ASM mass causing airway obstruction in inflammatory diseases such as asthma. Currently, there are no effective therapies to modulate ASM cell proliferation that contributes to ... Keywords Activator Protein-1; airway smooth muscle; retinoic acid; Airway Remodeling; Asthma; Protein Kinases; Transcription Factor AP-1; Tretinoin Download date 29/09/2021 14:19:54 Link to Item http://hdl.handle.net/10713/15769 Amy Elizabeth Defnet Contact Information: [email protected] Degree and Date to be Conferred: Ph.D. Pharmaceutical Sciences, May 2021 PROFESSIONAL OBJECTIVE My career objective is to work in academic institution where I can develop myself as an educator and researcher. My research employs a cross-disciplinary training regimen, including frequent opportunities for scientific/public speaking and inter-departmental engagement. In preparation for future teaching responsibilities, I have cultivated core pedagogical techniques through the JHU-UMB Collaborative Teaching Fellowship and Quality Matters Online Teaching Program. Additionally, participation in several societies and volunteer groups have helped cultivate my leadership and communication skills. EDUCATION University of Maryland, Baltimore 2016-present • Ph.D. Pharmaceutical Sciences, anticipated completion 2021 Fairleigh Dickinson University, Florham 2012-2016 • B.S. Biological Sciences with a Minor in Chemistry, 2016 RESEARCH Graduate Research Dr. Paul Shapiro and Dr. Maureen Kane, University of Maryland, Baltimore Fall 2016- Present This study hopes to overcome therapeutic limitations in asthma treatment that lead to bronchoconstriction and airway remodeling through evaluation of a novel function- selective ERK1/2 inhibitor and a RAR agonist. -

Identification, by Systematic RNA Sequencing, of Novel Candidate
Laboratory Investigation (2015) 95, 1077–1088 © 2015 USCAP, Inc All rights reserved 0023-6837/15 Identification, by systematic RNA sequencing, of novel candidate biomarkers and therapeutic targets in human soft tissue tumors Anne E Sarver1, Aaron L Sarver2, Venugopal Thayanithy1 and Subbaya Subramanian1 Human sarcomas comprise a heterogeneous group of more than 50 subtypes broadly classified into two groups: bone and soft tissue sarcomas. Such heterogeneity and their relative rarity have made them challenging targets for classification, biomarker identification, and development of improved treatment strategies. In this study, we used RNA sequencing to analyze 35 primary human tissue samples representing 13 different sarcoma subtypes, along with benign schwannoma, and normal bone and muscle tissues. For each sarcoma subtype, we detected unique messenger RNA (mRNA) expression signatures, which we further subjected to bioinformatic functional analysis, upstream regulatory analysis, and microRNA (miRNA) targeting analysis. We found that, for each sarcoma subtype, significantly upregulated genes and their deduced upstream regulators included not only previously implicated known players but also novel candidates not previously reported to be associated with sarcoma. For example, the schwannoma samples were characterized by high expression of not only the known associated proteins GFAP and GAP43 but also the novel player GJB6. Further, when we integrated our expression profiles with miRNA expression data from each sarcoma subtype, we were able to deduce potential key miRNA–gene regulator relationships for each. In the Ewing’s sarcoma and fibromatosis samples, two sarcomas where miR-182-5p is significantly downregulated, multiple predicted targets were significantly upregulated, including HMCN1, NKX2-2, SCNN1G, and SOX2. -

Family with Sequence Similarity 107: a Family of Stress Responsive Small Proteins with Diverse Functions in Cancer and the Nervous System (Review)
BIOMEDICAL REPORTS 2: 321-325, 2014 Family with sequence similarity 107: A family of stress responsive small proteins with diverse functions in cancer and the nervous system (Review) HIDEO NAKAJIMA1,2 and KEITA KOIZUMI3 1Department of Oncology, Ageo Central General Hospital, Ageo, Saitama 362-8588; 2Center for AIDS Research, Kumamoto University, Kumamoto 860-0811; 3Research Center for Child Mental Development, Kanazawa University, Kanazawa, Ishikawa 920-8640, Japan Received January 30, 2014; Accepted February 04, 2014 DOI: 10.3892/br.2014.243 Abstract. Under conditions of acute stress, rapid adaptation is Introduction crucial for maximizing biological survival. The responses to environmental stress are often complex, involving numerous Family with sequence similarity 107 (FAM107) members genes and integrating events at the cellular and organismal contain an N-terminal domain of unknown function levels. The heat shock proteins (HSPs) are a family of highly (DUF1151) that is conserved across species, including conserved proteins that play critical roles in maintaining cell mammalian, Xenopus, fish and Drosophila, with no homolo- homeostasis and protecting cells under chronic and acute gous matches to other functionally conserved domains (Fig. 1). stress conditions. The genes for these stress-responding This family includes several hypothetical eukaryotic proteins proteins are widely distributed in organisms, tissues and cells. with largely undetermined functions. Mammals have two HSPs participate in a variety of physiological processes and genes, FAM107A and FAM107B, which encode for proteins are associated with various types of disease. In this review, we of 144 and 131 amino acids (aa), respectively. The C-terminal focused on family with sequence similarity 107 (FAM107), a variable regions of FAM107 members have a coiled-coil novel unique protein family that exhibits functional similarity domain that has been identified in several nuclear proteins, with HSPs during the cellular stress response. -

Rare Allelic Variants in Meniere's Disease from Familial to Sporadic
PROGRAMA DE DOCTORADO EN BIOMEDICINA (B11.56.1) RARE ALLELIC VARIANTS IN MENIERE’S DISEASE FROM FAMILIAL TO SPORADIC CASES VARIANTES ALÉLICAS RARAS EN LA ENFERMEDAD DE MENIERE DE LOS CASOS FAMILIARES A LOS CASOS ESPORÁDICOS INTERNATIONAL PhD THESIS TESIS DOCTORAL CON MENCIÓN INTERNACIONAL ALVARO GALLEGO MARTINEZ February 2019 GRANADA Editor: Universidad de Granada. Tesis Doctorales Autor: Álvaro Gallego Martínez ISBN: 978-84-1306-154-2 URI: http://hdl.handle.net/10481/55469 Index Contents Index .............................................................................................................................................. 1 Grants and funding ........................................................................................................................ 3 Abstract ......................................................................................................................................... 5 Resumen ........................................................................................................................................ 6 Abbreviations ................................................................................................................................ 7 Introduction ................................................................................................................................... 9 Anatomy of the inner ear ........................................................................................................ 11 2. Meniere’s Disease .......................................................................................................... -

Induction of Therapeutic Tissue Tolerance Foxp3 Expression Is
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2012; 189:3947-3956; Prepublished online 17 from submission to initial decision 4 weeks from acceptance to publication September 2012; doi: 10.4049/jimmunol.1200449 http://www.jimmunol.org/content/189/8/3947 Foxp3 Expression Is Required for the Induction of Therapeutic Tissue Tolerance Frederico S. Regateiro, Ye Chen, Adrian R. Kendal, Robert Hilbrands, Elizabeth Adams, Stephen P. Cobbold, Jianbo Ma, Kristian G. Andersen, Alexander G. Betz, Mindy Zhang, Shruti Madhiwalla, Bruce Roberts, Herman Waldmann, Kathleen F. Nolan and Duncan Howie J Immunol cites 35 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/09/17/jimmunol.120044 9.DC1 This article http://www.jimmunol.org/content/189/8/3947.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021.