In Cancer Cells by Heat Shock Stimulation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Comprehensive Analysis of Mrna Expression Profiles in Head and Neck Cancer by Using Robust Rank Aggregation and Weighted Gene Coexpression Network Analysis
Hindawi BioMed Research International Volume 2020, Article ID 4908427, 21 pages https://doi.org/10.1155/2020/4908427 Research Article Comprehensive Analysis of mRNA Expression Profiles in Head and Neck Cancer by Using Robust Rank Aggregation and Weighted Gene Coexpression Network Analysis Zaizai Cao , Yinjie Ao , Yu Guo , and Shuihong Zhou Department of Otolaryngology, The First Affiliated Hospital, College of Medicine, Zhejiang University, Zhejiang Province 310003, China Correspondence should be addressed to Shuihong Zhou; [email protected] Received 3 June 2020; Revised 2 November 2020; Accepted 23 November 2020; Published 10 December 2020 Academic Editor: Hassan Dariushnejad Copyright © 2020 Zaizai Cao et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Head and neck squamous cell cancer (HNSCC) is the sixth most common cancer in the world; its pathogenic mechanism remains to be further clarified. Methods. Robust rank aggregation (RRA) analysis was utilized to identify the metasignature dysregulated genes, which were then used for potential drug prediction. Weighted gene coexpression network analysis (WGCNA) was performed on all metasignature genes to find hub genes. DNA methylation analysis, GSEA, functional annotation, and immunocyte infiltration analysis were then performed on hub genes to investigate their potential role in HNSCC. Result. A total of 862 metasignature genes were identified, and 6 potential drugs were selected based on these genes. Based on the result of WGCNA, six hub genes (ITM2A, GALNTL1, FAM107A, MFAP4, PGM5, and OGN) were selected (GS > 0:1, MM > 0:75,GSp value < 0.05, and MM p value < 0.05). -

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -
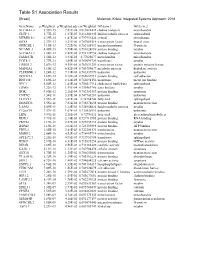
Table S1 Association Results
GeneName p.Weighted p.Weighted.adj cor.Weighted GO.term.1 GO.term.2 SLC44A1.2 3.55E-15 7.81E-08 0.819822422 choline transport mitochondria GLTP.1 5.77E-15 1.27E-07 0.816302445 lipid metabolic process sphingolipid MTMR10.1 6.39E-14 1.41E-06 0.797951424 cytosol phosphatase SOX8 2.37E-13 5.21E-06 0.787085514 transcription factor neural crest GPRC5B.1 4.19E-13 9.22E-06 0.782154937 integral membrane G-protein NCAM1.3 4.45E-13 9.79E-06 0.781624896 protein binding myelin SLC44A1.1 1.28E-12 2.82E-05 0.772132954 choline transport mitochondria FAM107B 1.56E-12 3.43E-05 0.77025677 mitochondria mitochondria UGT8.1 1.77E-12 3.89E-05 0.769099729 transferase myelin ERBB3.2 2.07E-12 4.55E-05 0.767631259 transcription factor protein tyrosine kinase MAN2A1 3.10E-12 6.82E-05 0.763759677 metabolic process hydrolase activity PLEKHH1.1 3.24E-12 7.13E-05 0.763337879 unknown unknown DOCK5.3 3.69E-12 8.12E-05 0.762087913 protein binding cell adhesion RNF130 3.69E-12 8.12E-05 0.762094156 membrane metal ion binding NPC1 6.50E-12 1.43E-04 0.756517114 cholesterol trafficking sphingolipid ERMN 7.22E-12 1.59E-04 0.755469786 actin binding myelin BOK 9.80E-12 2.16E-04 0.752383357 protein binding apoptosis CNTN2 1.54E-11 3.39E-04 0.747743281 unknown unknown ELOVL1 1.55E-11 3.41E-04 0.74764744 fatty acid sphingolipid DBNDD2 3.55E-11 7.81E-04 0.738878658 protein binding neuron projection LASS2 5.09E-11 1.12E-03 0.734954024 lipid metabolic process myelin C12orf34 7.57E-11 1.67E-03 0.730528911 unknown unknown LIPA 9.59E-11 2.11E-03 0.72786111 fatty acid glycerolipid metabolic -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype
Supplementary Information High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype Håkon Reikvam 1,2,*, Elise Aasebø 1, Annette K. Brenner 2, Sushma Bartaula-Brevik 1, Ida Sofie Grønningsæter 2, Rakel Brendsdal Forthun 2, Randi Hovland 3,4 and Øystein Bruserud 1,2 1 Department of Clinical Science, University of Bergen, 5020, Bergen, Norway 2 Department of Medicine, Haukeland University Hospital, 5021, Bergen, Norway 3 Department of Medical Genetics, Haukeland University Hospital, 5021, Bergen, Norway 4 Institute of Biomedicine, University of Bergen, 5020, Bergen, Norway * Correspondence: [email protected]; Tel.: +55-97-50-00 J. Clin. Med. 2019, 8, x 2 of 36 Figure S1. Mutational studies in a cohort of 71 AML patients. The figure shows the number of patients with the various mutations (upper), the number of mutations in for each patient (middle) and the number of main classes with mutation(s) in each patient (lower). 2 J. Clin. Med. 2019, 8, x; doi: www.mdpi.com/journal/jcm J. Clin. Med. 2019, 8, x 3 of 36 Figure S2. The immunophenotype of primary human AML cells derived from 62 unselected patients. The expression of the eight differentiation markers CD13, CD14, CD15, CD33, CD34, CD45, CD117 and HLA-DR was investigated for 62 of the 71 patients included in our present study. We performed an unsupervised hierarchical cluster analysis and identified four patient main clusters/patient subsets. The mutational profile for each f the 62 patients is also given (middle), no individual mutation of main class of mutations showed any significant association with any of the for differentiation marker clusters (middle). -
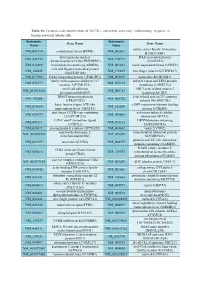
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Anti-FAM107A Antibody
D122337 Anti-FAM107A antibody Cat. No. D122337 Package 25 μl/100 μl/200 μl Storage -20°C, pH7.4 PBS, 0.05% NaN3, 40% Glycerol Product overview Description A n ti-FAM107A rabbit polyclonal antibody Applications ELISA, IHC Immunogen Fusion protein of human FAM107A Reactivity Human Content 0 .7 mg/ml Host species Rabbit Ig class Immunogen-specific rabbit IgG Purification Antigen affinity purification Target information Symbol FAM107A Full name family with sequence similarity 107, member A Synonyms DRR1; TU3A Swissprot O95990 Target Background FAM107B is a 131 amino acid protein that is encoded by a gene that maps to human chromosome 10, which contains over 800 genes and 135 million nucleotides, making up nearly 4.5% of the human genome. PTEN is an important tumor suppressor gene located on chromosome 10 and, when defective, causes a genetic predisposition to cancer development known as Cowden syndrome. The chromosome 10 encoded gene ERCC6 is important for DNA repair and is linked to Cockayne syndrome which is characterized by extreme photosensitivity and premature aging. Tetrahydrobiopterin deficiency and a number of syndromes involving defective skull and facial bone fusion are also linked to chromosoSme 10. angonAs with most trisomies, trisomy 10 is rarBie and is deloeteriouts. ech Applications Immunohistochemistry Predicted cell location: Nucleus and Cytoplasm Predicted cell location: Nucleus and Cytoplasm Positive control: Human liver cancer Positive control: Human colon cancer Recommended dilution: 50-200 Recommended dilution: 50-200 The image on the left is immunohistochemistry of paraffin-embedded The image on the left is immunohistochemistry of paraffin-embedded Human liver cancer tissue using D122337(FAM107A Antibody) at Human colon cancer tissue using D122337(FAM107A Antibody) at dilution 1/30, on the right is treated with fusion protein. -

LADON, a Natural Antisense Transcript of NODAL, Promotes Metastasis in Melanoma by Repressing NDRG1
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.09.032375; this version posted April 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. LADON, a natural antisense transcript of NODAL, promotes metastasis in melanoma by repressing NDRG1 Dutriaux Annie1, Diazzi Serena1, Caburet Sandrine2, Bresesti Chiara1, Hardouin Sylvie1, Deshayes Frédérique3, Collignon Jérôme1,* and Flagiello Domenico1* 1: Université de Paris, CNRS, Institut Jacques Monod, Regulation of Cell-Fate Specification team, Paris 75013, France 2: Université de Paris, CNRS, Institut Jacques Monod, Molecular Oncology and Ovarian Pathologies team, Paris 75013, France 3: Université de Paris, CNRS, Institut Jacques Monod, Morphogenesis, Homeostasis and Pathologies team, Paris 75013, France *Correspondence: [email protected]; [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.04.09.032375; this version posted April 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Summary The TGFb family member NODAL, primarily known for its role during embryonic development, has also been associated with tumor progression in a number of cancers. Some of the evidence supporting its involvement in melanoma has appeared contradictory, suggesting that NODAL in this context might rely on a non-canonical mode of signaling. To investigate this possibility we studied how a deletion of NODAL affected cell behavior in a metastatic melanoma cell line. The mutation does prevent melanoma cells from acquiring an invasive behavior. -

Supplementary Data
Supplemental figures Supplemental figure 1: Tumor sample selection. A total of 98 thymic tumor specimens were stored in Memorial Sloan-Kettering Cancer Center tumor banks during the study period. 64 cases corresponded to previously untreated tumors, which were resected upfront after diagnosis. Adjuvant treatment was delivered in 7 patients (radiotherapy in 4 cases, cyclophosphamide- doxorubicin-vincristine (CAV) chemotherapy in 3 cases). 34 tumors were resected after induction treatment, consisting of chemotherapy in 16 patients (cyclophosphamide-doxorubicin- cisplatin (CAP) in 11 cases, cisplatin-etoposide (PE) in 3 cases, cisplatin-etoposide-ifosfamide (VIP) in 1 case, and cisplatin-docetaxel in 1 case), in radiotherapy (45 Gy) in 1 patient, and in sequential chemoradiation (CAP followed by a 45 Gy-radiotherapy) in 1 patient. Among these 34 patients, 6 received adjuvant radiotherapy. 1 Supplemental Figure 2: Amino acid alignments of KIT H697 in the human protein and related orthologs, using (A) the Homologene database (exons 14 and 15), and (B) the UCSC Genome Browser database (exon 14). Residue H697 is highlighted with red boxes. Both alignments indicate that residue H697 is highly conserved. 2 Supplemental Figure 3: Direct comparison of the genomic profiles of thymic squamous cell carcinomas (n=7) and lung primary squamous cell carcinomas (n=6). (A) Unsupervised clustering analysis. Gains are indicated in red, and losses in green, by genomic position along the 22 chromosomes. (B) Genomic profiles and recurrent copy number alterations in thymic carcinomas and lung squamous cell carcinomas. Gains are indicated in red, and losses in blue. 3 Supplemental Methods Mutational profiling The exonic regions of interest (NCBI Human Genome Build 36.1) were broken into amplicons of 500 bp or less, and specific primers were designed using Primer 3 (on the World Wide Web for general users and for biologist programmers (see Supplemental Table 2) [1]. -

Heparanase Overexpression Induces Glucagon Resistance and Protects
Page 1 of 85 Diabetes Heparanase overexpression induces glucagon resistance and protects animals from chemically-induced diabetes Dahai Zhang1, Fulong Wang1, Nathaniel Lal1, Amy Pei-Ling Chiu1, Andrea Wan1, Jocelyn Jia1, Denise Bierende1, Stephane Flibotte1, Sunita Sinha1, Ali Asadi2, Xiaoke Hu2, Farnaz Taghizadeh2, Thomas Pulinilkunnil3, Corey Nislow1, Israel Vlodavsky4, James D. Johnson2, Timothy J. Kieffer2, Bahira Hussein1 and Brian Rodrigues1 1Faculty of Pharmaceutical Sciences, UBC, 2405 Wesbrook Mall, Vancouver, BC, Canada V6T 1Z3; 2Department of Cellular & Physiological Sciences, Life Sciences Institute, UBC, 2350 Health Sciences Mall, Vancouver, BC, Canada V6T 1Z3; 3Department of Biochemistry and Molecular Biology, Faculty of Medicine, Dalhousie University, 100 Tucker Park Road, Saint John, NB, Canada E2L 4L5; 4Cancer and Vascular Biology Research Center, Rappaport Faculty of Medicine, Technion, Haifa, Israel 31096 Running Title: Heparanase overexpression and the pancreatic islet Corresponding author: Dr. Brian Rodrigues Faculty of Pharmaceutical Sciences University of British Columbia, 2405 Wesbrook Mall, Vancouver, B.C., Canada V6T 1Z3 TEL: (604) 822-4758; FAX: (604) 822-3035 E-mail: [email protected] Key Words: Heparanase, heparan sulfate proteoglycan, glucose homeostasis, glucagon resistance, pancreatic islet, STZ Word Count: 4761 Total Number of DiabetesFigures: Publish 6 Ahead of Print, published online October 7, 2016 Diabetes Page 2 of 85 Abstract Heparanase, a protein with enzymatic and non-enzymatic properties, contributes towards disease progression and prevention. In the current study, a fortuitous observation in transgenic mice globally overexpressing heparanase (hep-tg) was the discovery of improved glucose homeostasis. We examined the mechanisms that contribute towards this improved glucose metabolism. Heparanase overexpression was associated with enhanced GSIS and hyperglucagonemia, in addition to changes in islet composition and structure. -

Rank Aggregation Via the Cross Entropy Algorithm
Finding cancer genes through meta-analysis of of the individual microarray studies were hybridized with the A®ymetrix GeneChip Human Genome HG-U133A and record expression levels for 22,283 Probe IDs. To make our ¯nal results meaningful and comprehensive at the same time, we decided to focus on microarray experiments that study di®erent types of cancer in humans. The goal of our meta-analysis is to identify genetic factors which are common across di®erent types and stages of cancer. For that purpose, we have selected 20 di®erent cancer-related microarray experiments which are included in the contest meta-dataset and have explicit cell type groupings necessary for detecting di®erentially expressed genes. Here, we list the selected experiment IDs along with the number of samples in each experiment in the parenthesis: E-MEXP-72 (20), E-MEXP-83 (22), E-MEXP-76 (17), E-MEXP-97 (24), E-MEXP-121 (30), E-MEXP-149 (20), E-MEXP-231 (58), E-MEXP-353 (96), E-TABM-26 (57), E-MEXP-669 (24), GSE4475 (221), GSE1456 (159), GSE5090 (17), GSE1420 (24), GSE1577 (29), GSE1729 (43), GSE2485 (18), GSE2603 (21), GSE3585 (12), GSE4127 (29). The total number of selected arrays is 941 (about 1/6 of the overall number of samples in the meta-dataset). One can refer to ArrayExpress database which provides public access to the microarray data from these experiments (http://www.ebi.ac.uk/arrayexpress/ ). 3 Methodology The proposed meta-analysis approach to microarray data is a two-step procedure: 1. Individual Analysis. By analyzing each microarray dataset individually, a set of \interesting" genes (top-50 Probe IDs) that exhibit the largest di®erences in terms of expression values between the groups is obtained for each dataset.