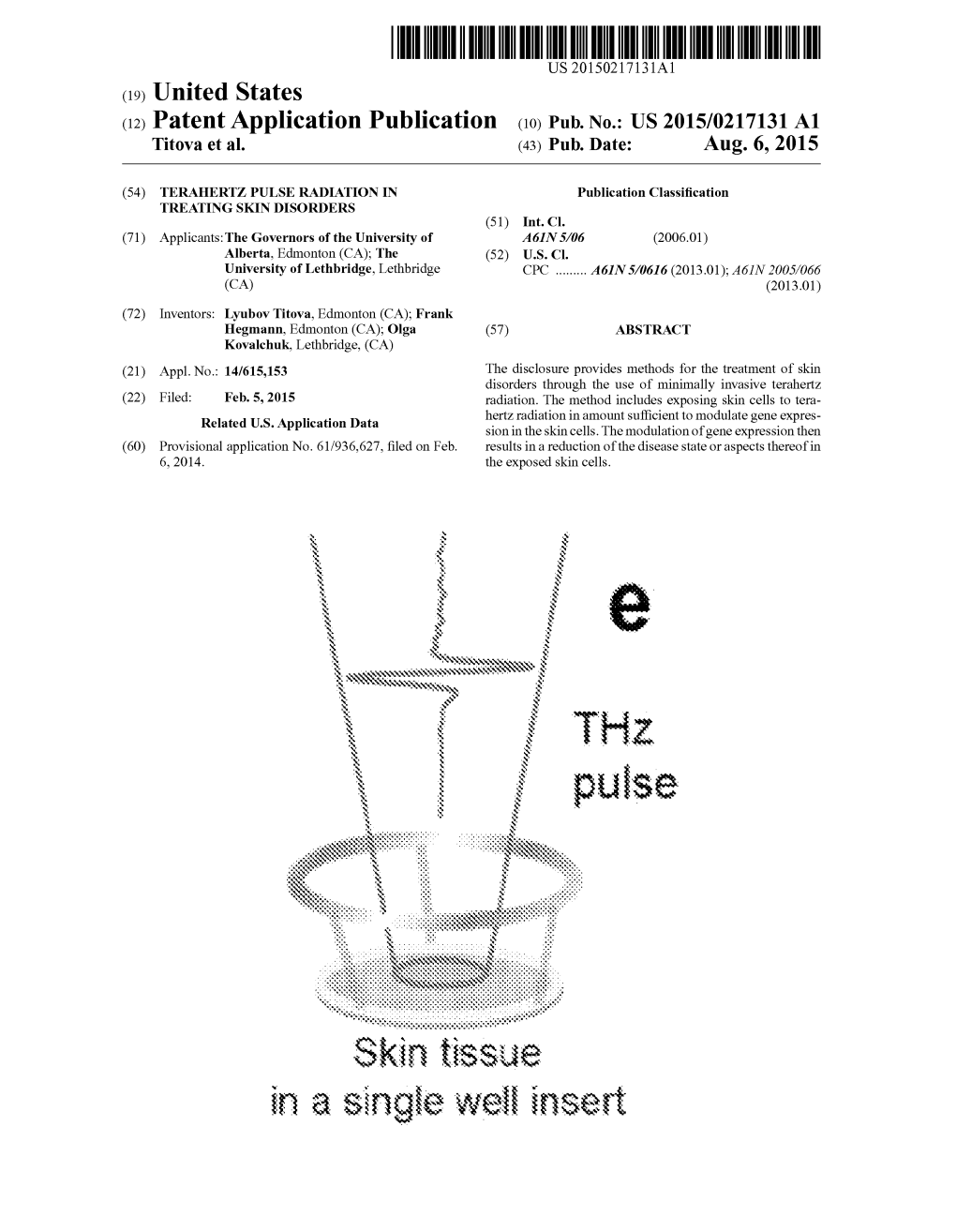
In a Single Wei Inset Patent Application Publication Aug
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

University of California, San Diego
UNIVERSITY OF CALIFORNIA, SAN DIEGO The post-terminal differentiation fate of RNAs revealed by next-generation sequencing A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Biomedical Sciences by Gloria Kuo Lefkowitz Committee in Charge: Professor Benjamin D. Yu, Chair Professor Richard Gallo Professor Bruce A. Hamilton Professor Miles F. Wilkinson Professor Eugene Yeo 2012 Copyright Gloria Kuo Lefkowitz, 2012 All rights reserved. The Dissertation of Gloria Kuo Lefkowitz is approved, and it is acceptable in quality and form for publication on microfilm and electronically: __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ Chair University of California, San Diego 2012 iii DEDICATION Ma and Ba, for your early indulgence and support. Matt and James, for choosing more practical callings. Roy, my love, for patiently sharing the ups and downs of this journey. iv EPIGRAPH It is foolish to tear one's hair in grief, as though sorrow would be made less by baldness. ~Cicero v TABLE OF CONTENTS Signature Page .............................................................................................................. iii Dedication .................................................................................................................... -

Small Cell Ovarian Carcinoma: Genomic Stability and Responsiveness to Therapeutics
Gamwell et al. Orphanet Journal of Rare Diseases 2013, 8:33 http://www.ojrd.com/content/8/1/33 RESEARCH Open Access Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics Lisa F Gamwell1,2, Karen Gambaro3, Maria Merziotis2, Colleen Crane2, Suzanna L Arcand4, Valerie Bourada1,2, Christopher Davis2, Jeremy A Squire6, David G Huntsman7,8, Patricia N Tonin3,4,5 and Barbara C Vanderhyden1,2* Abstract Background: The biology of small cell ovarian carcinoma of the hypercalcemic type (SCCOHT), which is a rare and aggressive form of ovarian cancer, is poorly understood. Tumourigenicity, in vitro growth characteristics, genetic and genomic anomalies, and sensitivity to standard and novel chemotherapeutic treatments were investigated in the unique SCCOHT cell line, BIN-67, to provide further insight in the biology of this rare type of ovarian cancer. Method: The tumourigenic potential of BIN-67 cells was determined and the tumours formed in a xenograft model was compared to human SCCOHT. DNA sequencing, spectral karyotyping and high density SNP array analysis was performed. The sensitivity of the BIN-67 cells to standard chemotherapeutic agents and to vesicular stomatitis virus (VSV) and the JX-594 vaccinia virus was tested. Results: BIN-67 cells were capable of forming spheroids in hanging drop cultures. When xenografted into immunodeficient mice, BIN-67 cells developed into tumours that reflected the hypercalcemia and histology of human SCCOHT, notably intense expression of WT-1 and vimentin, and lack of expression of inhibin. Somatic mutations in TP53 and the most common activating mutations in KRAS and BRAF were not found in BIN-67 cells by DNA sequencing. -
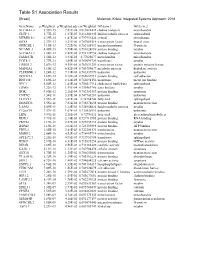
Table S1 Association Results
GeneName p.Weighted p.Weighted.adj cor.Weighted GO.term.1 GO.term.2 SLC44A1.2 3.55E-15 7.81E-08 0.819822422 choline transport mitochondria GLTP.1 5.77E-15 1.27E-07 0.816302445 lipid metabolic process sphingolipid MTMR10.1 6.39E-14 1.41E-06 0.797951424 cytosol phosphatase SOX8 2.37E-13 5.21E-06 0.787085514 transcription factor neural crest GPRC5B.1 4.19E-13 9.22E-06 0.782154937 integral membrane G-protein NCAM1.3 4.45E-13 9.79E-06 0.781624896 protein binding myelin SLC44A1.1 1.28E-12 2.82E-05 0.772132954 choline transport mitochondria FAM107B 1.56E-12 3.43E-05 0.77025677 mitochondria mitochondria UGT8.1 1.77E-12 3.89E-05 0.769099729 transferase myelin ERBB3.2 2.07E-12 4.55E-05 0.767631259 transcription factor protein tyrosine kinase MAN2A1 3.10E-12 6.82E-05 0.763759677 metabolic process hydrolase activity PLEKHH1.1 3.24E-12 7.13E-05 0.763337879 unknown unknown DOCK5.3 3.69E-12 8.12E-05 0.762087913 protein binding cell adhesion RNF130 3.69E-12 8.12E-05 0.762094156 membrane metal ion binding NPC1 6.50E-12 1.43E-04 0.756517114 cholesterol trafficking sphingolipid ERMN 7.22E-12 1.59E-04 0.755469786 actin binding myelin BOK 9.80E-12 2.16E-04 0.752383357 protein binding apoptosis CNTN2 1.54E-11 3.39E-04 0.747743281 unknown unknown ELOVL1 1.55E-11 3.41E-04 0.74764744 fatty acid sphingolipid DBNDD2 3.55E-11 7.81E-04 0.738878658 protein binding neuron projection LASS2 5.09E-11 1.12E-03 0.734954024 lipid metabolic process myelin C12orf34 7.57E-11 1.67E-03 0.730528911 unknown unknown LIPA 9.59E-11 2.11E-03 0.72786111 fatty acid glycerolipid metabolic -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-Like Mouse Models: Tracking the Role of the Hairless Gene
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 5-2006 Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene Yutao Liu University of Tennessee - Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Life Sciences Commons Recommended Citation Liu, Yutao, "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino- like Mouse Models: Tracking the Role of the Hairless Gene. " PhD diss., University of Tennessee, 2006. https://trace.tennessee.edu/utk_graddiss/1824 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Yutao Liu entitled "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the requirements for the degree of Doctor of Philosophy, with a major in Life Sciences. Brynn H. Voy, Major Professor We have read this dissertation and recommend its acceptance: Naima Moustaid-Moussa, Yisong Wang, Rogert Hettich Accepted for the Council: Carolyn R. -

Genome Sequence, Population History, and Pelage Genetics of the Endangered African Wild Dog (Lycaon Pictus) Michael G
Campana et al. BMC Genomics (2016) 17:1013 DOI 10.1186/s12864-016-3368-9 RESEARCH ARTICLE Open Access Genome sequence, population history, and pelage genetics of the endangered African wild dog (Lycaon pictus) Michael G. Campana1,2*, Lillian D. Parker1,2,3, Melissa T. R. Hawkins1,2,3, Hillary S. Young4, Kristofer M. Helgen2,3, Micaela Szykman Gunther5, Rosie Woodroffe6, Jesús E. Maldonado1,2 and Robert C. Fleischer1 Abstract Background: The African wild dog (Lycaon pictus) is an endangered African canid threatened by severe habitat fragmentation, human-wildlife conflict, and infectious disease. A highly specialized carnivore, it is distinguished by its social structure, dental morphology, absence of dewclaws, and colorful pelage. Results: We sequenced the genomes of two individuals from populations representing two distinct ecological histories (Laikipia County, Kenya and KwaZulu-Natal Province, South Africa). We reconstructed population demographic histories for the two individuals and scanned the genomes for evidence of selection. Conclusions: We show that the African wild dog has undergone at least two effective population size reductions in the last 1,000,000 years. We found evidence of Lycaon individual-specific regions of low diversity, suggestive of inbreeding or population-specific selection. Further research is needed to clarify whether these population reductions and low diversity regions are characteristic of the species as a whole. We documented positive selection on the Lycaon mitochondrial genome. Finally, we identified several candidate genes (ASIP, MITF, MLPH, PMEL) that may play a role in the characteristic Lycaon pelage. Keywords: Lycaon pictus, Genome, Population history, Selection, Pelage Background Primarily a hunter of antelopes, the African wild dog is a The African wild dog (Lycaon pictus) is an endangered highly distinct canine. -

Replace This with the Actual Title Using All Caps
UNDERSTANDING THE GENETICS UNDERLYING MASTITIS USING A MULTI-PRONGED APPROACH A Dissertation Presented to the Faculty of the Graduate School of Cornell University In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by Asha Marie Miles December 2019 © 2019 Asha Marie Miles UNDERSTANDING THE GENETICS UNDERLYING MASTITIS USING A MULTI-PRONGED APPROACH Asha Marie Miles, Ph. D. Cornell University 2019 This dissertation addresses deficiencies in the existing genetic characterization of mastitis due to granddaughter study designs and selection strategies based primarily on lactation average somatic cell score (SCS). Composite milk samples were collected across 6 sampling periods representing key lactation stages: 0-1 day in milk (DIM), 3- 5 DIM, 10-14 DIM, 50-60 DIM, 90-110 DIM, and 210-230 DIM. Cows were scored for front and rear teat length, width, end shape, and placement, fore udder attachment, udder cleft, udder depth, rear udder height, and rear udder width. Independent multivariable logistic regression models were used to generate odds ratios for elevated SCC (≥ 200,000 cells/ml) and farm-diagnosed clinical mastitis. Within our study cohort, loose fore udder attachment, flat teat ends, low rear udder height, and wide rear teats were associated with increased odds of mastitis. Principal component analysis was performed on these traits to create a single new phenotype describing mastitis susceptibility based on these high-risk phenotypes. Cows (N = 471) were genotyped on the Illumina BovineHD 777K SNP chip and considering all 14 traits of interest, a total of 56 genome-wide associations (GWA) were performed and 28 significantly associated quantitative trait loci (QTL) were identified. -

Autism Multiplex Family with 16P11.2P12.2 Microduplication Syndrome in Monozygotic Twins and Distal 16P11.2 Deletion in Their Brother
European Journal of Human Genetics (2012) 20, 540–546 & 2012 Macmillan Publishers Limited All rights reserved 1018-4813/12 www.nature.com/ejhg ARTICLE Autism multiplex family with 16p11.2p12.2 microduplication syndrome in monozygotic twins and distal 16p11.2 deletion in their brother Anne-Claude Tabet1,2,3,4, Marion Pilorge2,3,4, Richard Delorme5,6,Fre´de´rique Amsellem5,6, Jean-Marc Pinard7, Marion Leboyer6,8,9, Alain Verloes10, Brigitte Benzacken1,11,12 and Catalina Betancur*,2,3,4 The pericentromeric region of chromosome 16p is rich in segmental duplications that predispose to rearrangements through non-allelic homologous recombination. Several recurrent copy number variations have been described recently in chromosome 16p. 16p11.2 rearrangements (29.5–30.1 Mb) are associated with autism, intellectual disability (ID) and other neurodevelopmental disorders. Another recognizable but less common microdeletion syndrome in 16p11.2p12.2 (21.4 to 28.5–30.1 Mb) has been described in six individuals with ID, whereas apparently reciprocal duplications, studied by standard cytogenetic and fluorescence in situ hybridization techniques, have been reported in three patients with autism spectrum disorders. Here, we report a multiplex family with three boys affected with autism, including two monozygotic twins carrying a de novo 16p11.2p12.2 duplication of 8.95 Mb (21.28–30.23 Mb) characterized by single-nucleotide polymorphism array, encompassing both the 16p11.2 and 16p11.2p12.2 regions. The twins exhibited autism, severe ID, and dysmorphic features, including a triangular face, deep-set eyes, large and prominent nasal bridge, and tall, slender build. The eldest brother presented with autism, mild ID, early-onset obesity and normal craniofacial features, and carried a smaller, overlapping 16p11.2 microdeletion of 847 kb (28.40–29.25 Mb), inherited from his apparently healthy father. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

32-12012: Human Beta Defensin-3 Description Product
9853 Pacific Heights Blvd. Suite D. San Diego, CA 92121, USA Tel: 858-263-4982 Email: [email protected] 32-12012: Human Beta Defensin-3 Gene : DEFB103A Gene ID : 414325 Uniprot ID : P81534 Alternative Name : DEFB-3, Beta-defensin 3, Defensin, beta 103, Defensin-like protein Description Source: Genetically modified E.coli. Predicted MW: Monomer, 5.2 kDa (45 aa) Beta-Defensin 3 (BD-3), also known as DEFB-3, is a member of the defensin class of antimicrobial peptides. Beta defensins exert host defense responses against viruses, bacteria, and fungi through the binding and permeabilizing of microbial membranes. BD-3 expression is stimulated by interferon-gamma and is an important molecule during adaptive immunity. BD-3 functions to activate monocytes and mast cells, and has antibacterial functions towards Gram-negative and Gram-positive bacteria. Further, BD-3 blocks human immunodeficiency virus type 1 (HIV-1) replication through the downregulation of the HIV-1 co-receptor, CXCR4. Product Info Amount : 20 µg / 100 µg Purification : Reducing and Non-Reducing SDS PAGE at >= 95% Lyophilized from a sterile (0.2 micron) filtered aqueous solution containing 0.1% Trifluoroacetic Content : Acid (TFA) Sterile water at 0.1 mg/mL Storage condition : Store at -20°C Amino Acid : GIINTLQKYY CRVRGGRCAV LSCLPKEEQI GKCSTRGRKC CRRKK Application Note Endotoxin: Less than 0.1 ng/µg (1 IEU/µg) as determined by LAL test. Centrifuge vial before opening, Suspend the product by gently pipetting the above recommended solution down the sides of the vial. DO NOT VORTEX. Allow several minutes for complete reconstitution. For prolonged storage, dilute to working aliquots in a 0.1% BSA solution, store at -80°C and avoid repeat freeze thaws. -

Mapping DNA Structural Variation in Dogs
Downloaded from genome.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press Resource Mapping DNA structural variation in dogs Wei-Kang Chen,1,4 Joshua D. Swartz,1,4,5 Laura J. Rush,2 and Carlos E. Alvarez1,3,6 1Center for Molecular and Human Genetics, The Research Institute at Nationwide Children’s Hospital, Columbus, Ohio 43205, USA; 2Department of Veterinary Biosciences, The Ohio State University, Columbus, Ohio 43210, USA; 3Department of Pediatrics, The Ohio State University College of Medicine, Columbus, Ohio 43210, USA DNA structural variation (SV) comprises a major portion of genetic diversity, but its biological impact is unclear. We propose that the genetic history and extraordinary phenotypic variation of dogs make them an ideal mammal in which to study the effects of SV on biology and disease. The hundreds of existing dog breeds were created by selection of extreme morphological and behavioral traits. And along with those traits, each breed carries increased risk for different diseases. We used array CGH to create the first map of DNA copy number variation (CNV) or SV in dogs. The extent of this variation, and some of the gene classes affected, are similar to those of mice and humans. Most canine CNVs affect genes, including disease and candidate disease genes, and are thus likely to be functional. We identified many CNVs that may be breed or breed class specific. Cluster analysis of CNV regions showed that dog breeds tend to group according to breed classes. Our combined findings suggest many CNVs are (1) in linkage disequilibrium with flanking sequence, and (2) associated with breed-specific traits. -

Differential Epigenomic and Transcriptomic Responses in Subcutaneous Adipose Tissue Between Low and High Responders to Caloric Restriction1–3
Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction1–3 Luigi Bouchard, Re´mi Rabasa-Lhoret, May Faraj, Marie-E`ve Lavoie, Jonathan Mill, Louis Pe´russe, and Marie-Claude Vohl ABSTRACT weight loss responses to caloric restriction show considerable Background: Caloric restriction is recommended for the treatment interindividual variability (7). Studies of genetically identical of obesity, but it is generally characterized by large interindividual monozygotic twins have been particularly useful in disentangling variability in responses. The factors affecting the magnitude of the role of environmental and heritable factors in determining the weight loss remain poorly understood. Epigenetic factors (ie, heri- degree of weight loss. It has been shown that within-pair changes table but reversible changes to genomic function that regulate gene in body fat variability after a caloric deficit is significantly lower expression independently of DNA sequence) may explain some of than between-pair variability, which suggests that genetic factors the interindividual variability seen in weight-loss responses. have an important influence on an individual’s response to caloric Objective: The objective was to determine whether epigenetics and deficit (8, 9). However, the concordance between twin pairs was gene expression changes may play a role in weight-loss responsiveness. not complete, which suggests that environmental factors or other Design: Overweight/obese postmenopausal women were recruited DNA sequence–independent mechanisms may be involved. for a standard 6-mo caloric restriction intervention. Abdominal sub- It has been suggested that monozygotic twin discordance for cutaneous adipose tissue biopsy samples were collected before (n = complex traits such as body weight could be accounted for by 14) and after (n = 14) intervention, and the epigenomic and tran- epigenetic factors (10, 11).