Role of Microrna in Early Life Placental Programming of Insulin Resistance and Metabolic Health
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
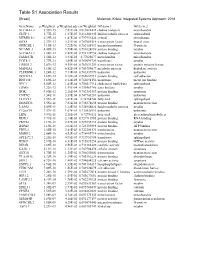
Table S1 Association Results
GeneName p.Weighted p.Weighted.adj cor.Weighted GO.term.1 GO.term.2 SLC44A1.2 3.55E-15 7.81E-08 0.819822422 choline transport mitochondria GLTP.1 5.77E-15 1.27E-07 0.816302445 lipid metabolic process sphingolipid MTMR10.1 6.39E-14 1.41E-06 0.797951424 cytosol phosphatase SOX8 2.37E-13 5.21E-06 0.787085514 transcription factor neural crest GPRC5B.1 4.19E-13 9.22E-06 0.782154937 integral membrane G-protein NCAM1.3 4.45E-13 9.79E-06 0.781624896 protein binding myelin SLC44A1.1 1.28E-12 2.82E-05 0.772132954 choline transport mitochondria FAM107B 1.56E-12 3.43E-05 0.77025677 mitochondria mitochondria UGT8.1 1.77E-12 3.89E-05 0.769099729 transferase myelin ERBB3.2 2.07E-12 4.55E-05 0.767631259 transcription factor protein tyrosine kinase MAN2A1 3.10E-12 6.82E-05 0.763759677 metabolic process hydrolase activity PLEKHH1.1 3.24E-12 7.13E-05 0.763337879 unknown unknown DOCK5.3 3.69E-12 8.12E-05 0.762087913 protein binding cell adhesion RNF130 3.69E-12 8.12E-05 0.762094156 membrane metal ion binding NPC1 6.50E-12 1.43E-04 0.756517114 cholesterol trafficking sphingolipid ERMN 7.22E-12 1.59E-04 0.755469786 actin binding myelin BOK 9.80E-12 2.16E-04 0.752383357 protein binding apoptosis CNTN2 1.54E-11 3.39E-04 0.747743281 unknown unknown ELOVL1 1.55E-11 3.41E-04 0.74764744 fatty acid sphingolipid DBNDD2 3.55E-11 7.81E-04 0.738878658 protein binding neuron projection LASS2 5.09E-11 1.12E-03 0.734954024 lipid metabolic process myelin C12orf34 7.57E-11 1.67E-03 0.730528911 unknown unknown LIPA 9.59E-11 2.11E-03 0.72786111 fatty acid glycerolipid metabolic -

The Title of the Article
Mechanism-Anchored Profiling Derived from Epigenetic Networks Predicts Outcome in Acute Lymphoblastic Leukemia Xinan Yang, PhD1, Yong Huang, MD1, James L Chen, MD1, Jianming Xie, MSc2, Xiao Sun, PhD2, Yves A Lussier, MD1,3,4§ 1Center for Biomedical Informatics and Section of Genetic Medicine, Department of Medicine, The University of Chicago, Chicago, IL 60637 USA 2State Key Laboratory of Bioelectronics, Southeast University, 210096 Nanjing, P.R.China 3The University of Chicago Cancer Research Center, and The Ludwig Center for Metastasis Research, The University of Chicago, Chicago, IL 60637 USA 4The Institute for Genomics and Systems Biology, and the Computational Institute, The University of Chicago, Chicago, IL 60637 USA §Corresponding author Email addresses: XY: [email protected] YH: [email protected] JC: [email protected] JX: [email protected] XS: [email protected] YL: [email protected] - 1 - Abstract Background Current outcome predictors based on “molecular profiling” rely on gene lists selected without consideration for their molecular mechanisms. This study was designed to demonstrate that we could learn about genes related to a specific mechanism and further use this knowledge to predict outcome in patients – a paradigm shift towards accurate “mechanism-anchored profiling”. We propose a novel algorithm, PGnet, which predicts a tripartite mechanism-anchored network associated to epigenetic regulation consisting of phenotypes, genes and mechanisms. Genes termed as GEMs in this network meet all of the following criteria: (i) they are co-expressed with genes known to be involved in the biological mechanism of interest, (ii) they are also differentially expressed between distinct phenotypes relevant to the study, and (iii) as a biomodule, genes correlate with both the mechanism and the phenotype. -

Proteomic Identification of the Transcription Factors Ikaros And
European School of Molecular Medicine (SEMM) University of Milan and University of Naples “Federico II” PhD degree in Systems Medicine (curriculum in Molecular Oncology) Settore disciplinare: BIO/11 Proteomic identification of the transcription factors Ikaros and Aiolos as new Myc interactors on chromatin Chiara Veronica Locarno Matricola: R10755 Center for Genomic Science IIT@SEMM, Milan Supervisor: Bruno Amati, PhD IEO, Milan Added Supervisor: Arianna Sabò, PhD IEO, Milan Academic year 2017-2018 Table of contents List of abbreviations ........................................................................................................... 4 List of figures ....................................................................................................................... 8 List of tables ....................................................................................................................... 11 Abstract .............................................................................................................................. 12 1. INTRODUCTION ......................................................................................................... 13 1.1 Myc ........................................................................................................................................ 13 1.1.1 Myc discovery and structure ........................................................................................... 13 1.1.2. Role of Myc in physiological and pathological conditions ........................................... -

Broad Poster Vivek
A novel computational method for finding regions with copy number abnormalities in cancer cells Vivek, Manuel Garber, and Mike Zody Broad Institute of MIT and Harvard, Cambridge, MA, USA Introduction Results Cancer can result from the over expression of oncogenes, genes which control and regulate cell growth. Sometimes oncogenes increase in 1 2 3 activity due to a specific genetic mutation called a translocation (Fig 1). SMAD4 – a gene known to be deleted in pancreatic COX10 – a gene deleted in cytochrome c oxidase AK001392 – a hereditary prostate cancer protein This translocation allows the oncogene to remain as active as its paired carcinoma deficiency, known to be related to cell proliferation gene. Amplification of this mutation can occur, thereby creating the proper conditions for uncontrolled cell growth; consequently, each Results from Analysis Program Results from Analysis Program Results from Analysis Program component of the translocation will amplify in similar quantities. In this mutation, the chromosomal region containing the oncogene displaces to Region 1 Region 2 R2 Region 1 Region 2 R2 Region 1 Region 2 R2 a region on another chromosome containing a gene that is expressed Chr18:47044749-47311978 Chr17:13930739-14654741 0.499070821478475 Chr17:13930739-14654741 Chr18:26861790-27072166 0.47355172850856 Chr17:12542326-13930738 Chr8:1789292-1801984 0.406208680312004 frequently. Actual region containing gene Actual region containing gene Actual region containing gene chr18: 45,842,214 - 48,514,513 chr17: 13,966,862 - 14,068,461 chr17: 12,542,326 - 13,930,738 Fig 1. Two chromosomal regions (abcdef and ghijk) are translocating to create two new regions (abckl and ghijedf). -

Download on 20
bioRxiv preprint doi: https://doi.org/10.1101/850776; this version posted January 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Intramembrane protease RHBDL4 cleaves oligosaccharyltransferase subunits to target them for ER-associated degradation Julia D. Knopf1, Nina Landscheidt1, Cassandra L. Pegg2, Benjamin L. Schulz2, Nathalie Kühnle1, Chao-Wei Chao1, Simon Huck1 and Marius K. Lemberg1, # 1Centre for Molecular Biology of Heidelberg University (ZMBH), DKFZ-ZMBH Alliance, 69120 Heidelberg, Germany. 2School of Chemistry and Molecular Biosciences, ARC Training Centre for Biopharmaceutical Innovation, The University of Queensland, St Lucia QLD 4072, Australia. #Corresponding author: [email protected] Running title: RHBDL4 triggers ERAD of OST subunits Key words: Rhomboid serine protease, Rhbdd1, ubiquitin-dependent proteolysis, post- translational protein abundance control, N-linked glycosylation. Abbreviations ERAD, ER-associated degradation; OST, oligosacharyltransferase; TM, transmembrane; UIM, ubiquitin-interacting motif. Abstract The Endoplasmic Reticulum (ER)-resident intramembrane rhomboid protease RHBDL4 generates metastable protein fragments and together with the ER-associated degradation (ERAD) machinery provides a clearance mechanism for aberrant and surplus proteins. However, the endogenous substrate spectrum and with that the role of RHBDL4 in physiological ERAD is mainly unknown. Here, we use a substrate trapping approach in combination with quantitative proteomics to identify physiological RHBDL4 substrates. This revealed oligosacharyltransferase (OST) complex subunits such as the catalytic active subunit STT3A as substrates for the RHBDL4-dependent ERAD pathway. RHBDL4-catalyzed cleavage inactivates OST subunits by triggering dislocation into the cytoplasm and subsequent proteasomal degradation. -

TASOR Is a Pseudo-PARP That Directs HUSH Complex Assembly and Epigenetic Transposon Control
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.09.974832; this version posted March 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control Christopher H. Douse1,‡, Iva A. Tchasovnikarova2,‡, Richard T. Timms2,‡, Anna V. Protasio2, Marta Seczynska2, Daniil M. Prigozhin1, Anna Albecka1,2, Jane Wagstaff3, James C. Williamson2, Stefan M.V. Freund3, Paul J. Lehner2*, Yorgo Modis1,2* 1 Molecular Immunity Unit, Department of Medicine, University of Cambridge, MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge, CB2 0QH, UK 2 Cambridge Institute of Therapeutic Immunology & Infectious Disease (CITIID), Department of Medicine, University of Cambridge, Cambridge CB2 0AW, UK 3 Structural Studies Division, MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge, CB2 0QH, UK Current addresses: Department of Experimental Medical Science, Lund University, Sölvegatan 19, Lund, Sweden (C.H.D.); The Gurdon Institute, Tennis Court Road, Cambridge, UK (I.A.T.); Department of Pathology, Tennis Court Road, Cambridge, UK (A.V.P.); Molecular Biophysics and Integrated Bioimaging Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA (D.M.P.) ‡These authors contributed equally *co-corresponding authors Keywords: Transcriptional repression; epigenetic silencing; antiretroviral response; genome stability; histone H3 lysine 9 methylation (H3K9me3); transposable element (TE); long interspersed nuclear element-1 (LINE-1); poly-ADP ribose polymerase (PARP); RNA-binding protein; RNA-induced transcriptional silencing; CUT&RUN; CUT&Tag; genome profiling 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.03.09.974832; this version posted March 11, 2020. -

1 ICR-Geneset Gene List
ICR-geneset Gene List. IMAGE ID UniGene Locus Name Cluster 20115 Hs.62185 SLC9A6 solute carrier family 9 (sodium/hydrogen exchanger), isoform 6 21738 21899 Hs.78353 SRPK2 SFRS protein kinase 2 21908 Hs.79133 CDH8 cadherin 8, type 2 22040 Hs.151738 MMP9 matrix metalloproteinase 9 (gelatinase B, 92kD gelatinase, 92kD type IV collagenase) 22411 Hs.183 FY Duffy blood group 22731 Hs.1787 PHRET1 PH domain containing protein in retina 1 22859 Hs.356487 ESTs 22883 Hs.150926 FPGT fucose-1-phosphate guanylyltransferase 22918 Hs.346868 EBNA1BP2 EBNA1 binding protein 2 23012 Hs.158205 BLZF1 basic leucine zipper nuclear factor 1 (JEM-1) 23073 Hs.284244 FGF2 fibroblast growth factor 2 (basic) 23173 Hs.151051 MAPK10 mitogen-activated protein kinase 10 23185 Hs.289114 TNC tenascin C (hexabrachion) 23282 Hs.8024 IK IK cytokine, down-regulator of HLA II 23353 23431 Hs.50421 RB1CC1 RB1-inducible coiled-coil 1 23514 23548 Hs.71848 Human clone 23548 mRNA sequence 23629 Hs.135587 Human clone 23629 mRNA sequence 23658 Hs.265855 SETMAR SET domain and mariner transposase fusion gene 23676 Hs.100841 Homo sapiens clone 23676 mRNA sequence 23772 Hs.78788 LZTR1 leucine-zipper-like transcriptional regulator, 1 23776 Hs.75438 QDPR quinoid dihydropteridine reductase 23804 Hs.343586 ZFP36 zinc finger protein 36, C3H type, homolog (mouse) 23831 Hs.155247 ALDOC aldolase C, fructose-bisphosphate 23878 Hs.99902 OPCML opioid binding protein/cell adhesion molecule-like 23903 Hs.12526 Homo sapiens clone 23903 mRNA sequence 23932 Hs.368063 Human clone 23932 mRNA sequence 24004 -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Exploring Prostate Cancer Genome Reveals Simultaneous Losses of PTEN, FAS and PAPSS2 in Patients with PSA Recurrence After Radical Prostatectomy
Int. J. Mol. Sci. 2015, 16, 3856-3869; doi:10.3390/ijms16023856 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Exploring Prostate Cancer Genome Reveals Simultaneous Losses of PTEN, FAS and PAPSS2 in Patients with PSA Recurrence after Radical Prostatectomy Chinyere Ibeawuchi 1, Hartmut Schmidt 2, Reinhard Voss 3, Ulf Titze 4, Mahmoud Abbas 5, Joerg Neumann 6, Elke Eltze 7, Agnes Marije Hoogland 8, Guido Jenster 9, Burkhard Brandt 10 and Axel Semjonow 1,* 1 Prostate Center, Department of Urology, University Hospital Muenster, Albert-Schweitzer-Campus 1, Gebaeude 1A, Muenster D-48149, Germany; E-Mail: [email protected] 2 Center for Laboratory Medicine, University Hospital Muenster, Albert-Schweitzer-Campus 1, Gebaeude 1A, Muenster D-48149, Germany; E-Mail: [email protected] 3 Interdisciplinary Center for Clinical Research, University of Muenster, Albert-Schweitzer-Campus 1, Gebaeude D3, Domagkstrasse 3, Muenster D-48149, Germany; E-Mail: [email protected] 4 Pathology, Lippe Hospital Detmold, Röntgenstrasse 18, Detmold D-32756, Germany; E-Mail: [email protected] 5 Institute of Pathology, Mathias-Spital-Rheine, Frankenburg Street 31, Rheine D-48431, Germany; E-Mail: [email protected] 6 Institute of Pathology, Klinikum Osnabrueck, Am Finkenhuegel 1, Osnabrueck D-49076, Germany; E-Mail: [email protected] 7 Institute of Pathology, Saarbrücken-Rastpfuhl, Rheinstrasse 2, Saarbrücken D-66113, Germany; E-Mail: [email protected] 8 Department -

Download The
PROBING THE INTERACTION OF ASPERGILLUS FUMIGATUS CONIDIA AND HUMAN AIRWAY EPITHELIAL CELLS BY TRANSCRIPTIONAL PROFILING IN BOTH SPECIES by POL GOMEZ B.Sc., The University of British Columbia, 2002 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE in THE FACULTY OF GRADUATE STUDIES (Experimental Medicine) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2010 © Pol Gomez, 2010 ABSTRACT The cells of the airway epithelium play critical roles in host defense to inhaled irritants, and in asthma pathogenesis. These cells are constantly exposed to environmental factors, including the conidia of the ubiquitous mould Aspergillus fumigatus, which are small enough to reach the alveoli. A. fumigatus is associated with a spectrum of diseases ranging from asthma and allergic bronchopulmonary aspergillosis to aspergilloma and invasive aspergillosis. Airway epithelial cells have been shown to internalize A. fumigatus conidia in vitro, but the implications of this process for pathogenesis remain unclear. We have developed a cell culture model for this interaction using the human bronchial epithelium cell line 16HBE and a transgenic A. fumigatus strain expressing green fluorescent protein (GFP). Immunofluorescent staining and nystatin protection assays indicated that cells internalized upwards of 50% of bound conidia. Using fluorescence-activated cell sorting (FACS), cells directly interacting with conidia and cells not associated with any conidia were sorted into separate samples, with an overall accuracy of 75%. Genome-wide transcriptional profiling using microarrays revealed significant responses of 16HBE cells and conidia to each other. Significant changes in gene expression were identified between cells and conidia incubated alone versus together, as well as between GFP positive and negative sorted cells. -

Plenary and Platform Abstracts
American Society of Human Genetics 68th Annual Meeting PLENARY AND PLATFORM ABSTRACTS Abstract #'s Tuesday, October 16, 5:30-6:50 pm: 4. Featured Plenary Abstract Session I Hall C #1-#4 Wednesday, October 17, 9:00-10:00 am, Concurrent Platform Session A: 6. Variant Insights from Large Population Datasets Ballroom 20A #5-#8 7. GWAS in Combined Cancer Phenotypes Ballroom 20BC #9-#12 8. Genome-wide Epigenomics and Non-coding Variants Ballroom 20D #13-#16 9. Clonal Mosaicism in Cancer, Alzheimer's Disease, and Healthy Room 6A #17-#20 Tissue 10. Genetics of Behavioral Traits and Diseases Room 6B #21-#24 11. New Frontiers in Computational Genomics Room 6C #25-#28 12. Bone and Muscle: Identifying Causal Genes Room 6D #29-#32 13. Precision Medicine Initiatives: Outcomes and Lessons Learned Room 6E #33-#36 14. Environmental Exposures in Human Traits Room 6F #37-#40 Wednesday, October 17, 4:15-5:45 pm, Concurrent Platform Session B: 24. Variant Interpretation Practices and Resources Ballroom 20A #41-#46 25. Integrated Variant Analysis in Cancer Genomics Ballroom 20BC #47-#52 26. Gene Discovery and Functional Models of Neurological Disorders Ballroom 20D #53-#58 27. Whole Exome and Whole Genome Associations Room 6A #59-#64 28. Sequencing-based Diagnostics for Newborns and Infants Room 6B #65-#70 29. Omics Studies in Alzheimer's Disease Room 6C #71-#76 30. Cardiac, Valvular, and Vascular Disorders Room 6D #77-#82 31. Natural Selection and Human Phenotypes Room 6E #83-#88 32. Genetics of Cardiometabolic Traits Room 6F #89-#94 Wednesday, October 17, 6:00-7:00 pm, Concurrent Platform Session C: 33. -

Ten Commandments for a Good Scientist
Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds Ana María Sotoca Covaleda Wageningen 2010 Thesis committee Thesis supervisors Prof. dr. ir. Ivonne M.C.M. Rietjens Professor of Toxicology Wageningen University Prof. dr. Albertinka J. Murk Personal chair at the sub-department of Toxicology Wageningen University Thesis co-supervisor Dr. ir. Jacques J.M. Vervoort Associate professor at the Laboratory of Biochemistry Wageningen University Other members Prof. dr. Michael R. Muller, Wageningen University Prof. dr. ir. Huub F.J. Savelkoul, Wageningen University Prof. dr. Everardus J. van Zoelen, Radboud University Nijmegen Dr. ir. Toine F.H. Bovee, RIKILT, Wageningen This research was conducted under the auspices of the Graduate School VLAG Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds Ana María Sotoca Covaleda Thesis submitted in fulfillment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. dr. M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Tuesday 14 September 2010 at 4 p.m. in the Aula Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds. Ana María Sotoca Covaleda Thesis Wageningen University, Wageningen, The Netherlands, 2010, With references, and with summary in Dutch. ISBN: 978-90-8585-707-5 “Caminante no hay camino, se hace camino al andar. Al andar se hace camino, y al volver la vista atrás se ve la senda que nunca se ha de volver a pisar” - Antonio Machado – A mi madre.